2YNI
 
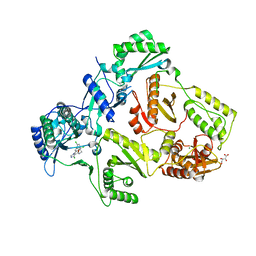 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK952 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNF
 
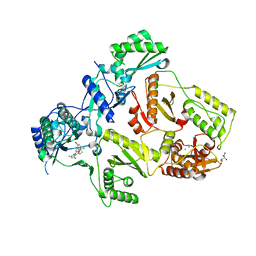 | | HIV-1 Reverse Transcriptase Y188L mutant in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNG
 
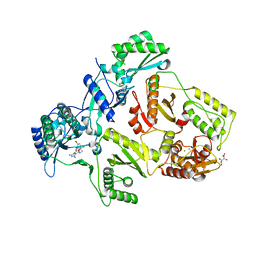 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK560 | | Descriptor: | 2-azanyl-N-[[4-bromanyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-4-chloranyl-1H-imidazole-5-carboxamide, MAGNESIUM ION, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2YNH
 
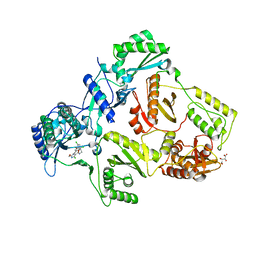 | | HIV-1 Reverse Transcriptase in complex with inhibitor GSK500 | | Descriptor: | 4-chloranyl-N-[[4-chloranyl-3-(3-chloranyl-5-cyano-phenoxy)-2-fluoranyl-phenyl]methyl]-2-(hydroxymethyl)-1H-imidazole-5-carboxamide, D(-)-TARTARIC ACID, P51 RT, ... | | Authors: | Chong, P, Sebahar, P, Youngman, M, Garrido, D, Zhang, H, Stewart, E.L, Nolte, R.T, Wang, L, Ferris, R.G, Edelstein, M, Weaver, K, Mathis, A, Peat, A. | | Deposit date: | 2012-10-14 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational Design of Potent Non-Nucleoside Inhibitors of HIV-1 Reverse Transcriptase.
J.Med.Chem., 55, 2012
|
|
2DJY
 
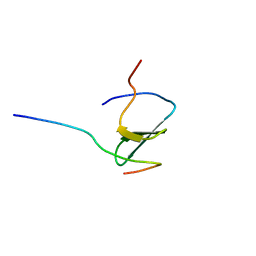 | | Solution structure of Smurf2 WW3 domain-Smad7 PY peptide complex | | Descriptor: | Mothers against decapentaplegic homolog 7, Smad ubiquitination regulatory factor 2 | | Authors: | Chong, P.A, Lin, H, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2006-04-06 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Expanded WW Domain Recognition Motif Revealed by the Interaction between Smad7 and the E3 Ubiquitin Ligase Smurf2.
J.Biol.Chem., 281, 2006
|
|
5NOP
 
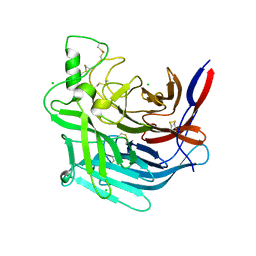 | | Structure of Mojiang virus attachment glycoprotein | | Descriptor: | Attachment glycoprotein, CHLORIDE ION | | Authors: | Rissanen, I.R, Ahmed, A.A, Beaty, S, Azarm, K, Hong, P, Nambulli, S, Duprex, P.W, Lee, B, Bowden, T.A. | | Deposit date: | 2017-04-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Idiosyncratic Mojiang virus attachment glycoprotein directs a host-cell entry pathway distinct from genetically related henipaviruses.
Nat Commun, 8, 2017
|
|
8K4Q
 
 | |
4B3K
 
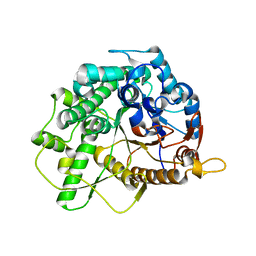 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4B3L
 
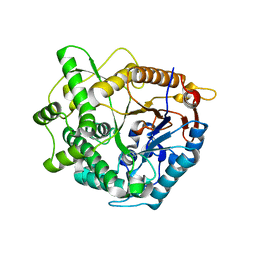 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1PYN
 
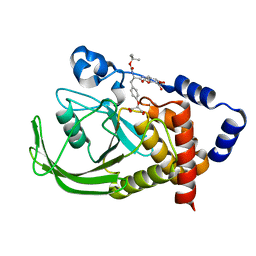 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1QAN
 
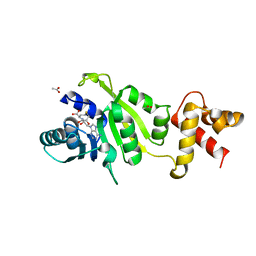 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ACETATE ION, ERMC' METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-26 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAO
 
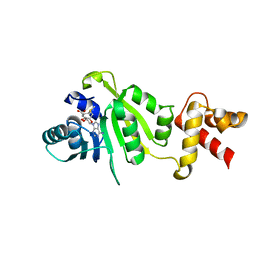 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ERMC' METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-26 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAM
 
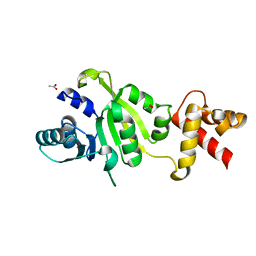 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ACETATE ION, ERMC' METHYLTRANSFERASE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-25 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAQ
 
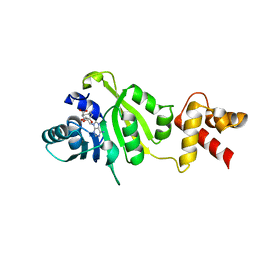 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ERMC' RRNA METHYLTRANSFERASE, SINEFUNGIN | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-28 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
7E53
 
 | |
2XVC
 
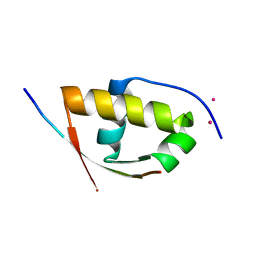 | | Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division | | Descriptor: | CADMIUM ION, CDVA, SSO0911, ... | | Authors: | Samson, R.Y, Obita, T, Hodgson, B, Shaw, M.K, Chong, P.L, Williams, R.L, Bell, S.D. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular and Structural Basis of Escrt-III Recruitment to Membranes During Archaeal Cell Division.
Mol.Cell, 41, 2011
|
|
2BL9
 
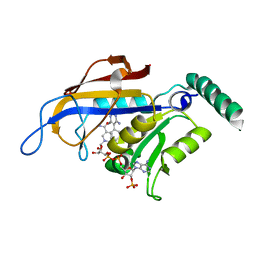 | | X-ray crystal structure of Plasmodium vivax dihydrofolate reductase in complex with pyrimethamine and its derivative | | Descriptor: | 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-02 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLC
 
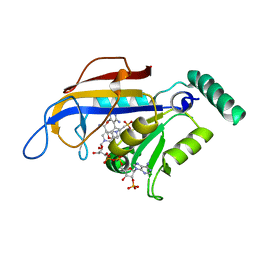 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with des-chloropyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLB
 
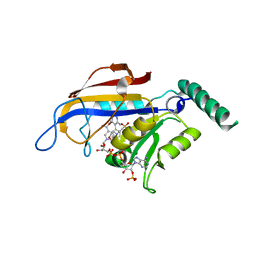 | | X-ray crystal structure of Plasmodium vivax dihydrofolate reductase in complex with pyrimethamine and its derivative | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-ETHYL-5-PHENYLPYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-03 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLA
 
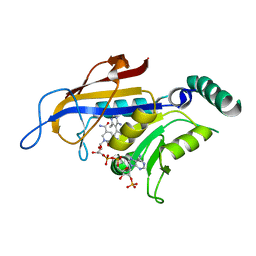 | | SP21 double mutant P. vivax Dihydrofolate reductase in complex with pyrimethamine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE, ... | | Authors: | Kongsaeree, P, Khongsuk, P, Leartsakulpanich, U, Chitnumsub, P, Tarnchompoo, B, Walkinshaw, M.D, Yuthavong, Y. | | Deposit date: | 2005-03-02 | | Release date: | 2005-09-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Plasmodium Vivax: Pyrimethamine Displacement Linked with Mutation-Induced Resistance.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1YUB
 
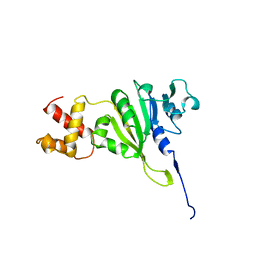 | | SOLUTION STRUCTURE OF AN RRNA METHYLTRANSFERASE (ERMAM) THAT CONFERS MACROLIDE-LINCOSAMIDE-STREPTOGRAMIN ANTIBIOTIC RESISTANCE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | RRNA METHYLTRANSFERASE | | Authors: | Yu, L, Petros, A.M, Schnuchel, A, Zhong, P, Severin, J.M, Walter, K, Holzman, T.F, Fesik, S.W. | | Deposit date: | 1997-03-04 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance.
Nat.Struct.Biol., 4, 1997
|
|
2LC4
 
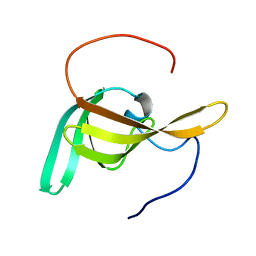 | | Solution Structure of PilP from Pseudomonas aeruginosa | | Descriptor: | PilP protein | | Authors: | Howell, P, Tammam, S, Chong, P, Forman-Kay, J.D. | | Deposit date: | 2011-04-22 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the PilN, PilO and PilP type IVa pilus subcomplex.
Mol.Microbiol., 82, 2011
|
|
6KLS
 
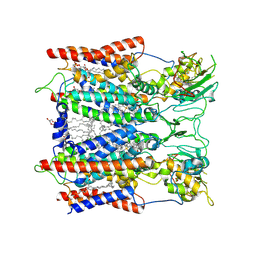 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, Cytochrome c, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6ITP
 
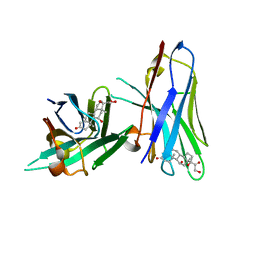 | | Crystal structure of cortisol complexed with its nanobody at pH 3.5 | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol camelid antibody | | Authors: | Ding, Y, Ding, L.L, Wang, Z.Y, Zhong, P.Y. | | Deposit date: | 2018-11-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Structural insights into the mechanism of single domain VHH antibody binding to cortisol.
Febs Lett., 593, 2019
|
|
6KLV
 
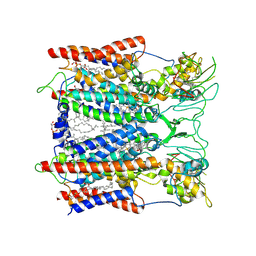 | | Hyperthermophilic respiratory Complex III | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione, ANTIMYCIN, ... | | Authors: | Fei, S, Hartmut, M, Yun, Z, Guohong, P, Guoliang, Z, Hui, Z, Shuangbo, Z, Xiaoyun, P, Yan, Z. | | Deposit date: | 2019-07-30 | | Release date: | 2020-05-13 | | Last modified: | 2021-04-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 3.3 angstrom -Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
