2N9W
 
 | | Solution NMR Structure of the membrane localization domain from the Ras/Rap1-specific endopeptidase (RRSP) of the Vibrio vulnificus multifunctional autoprocessing repeats-in-toxins (MARTX) toxin | | Descriptor: | RTX toxin | | Authors: | Hisao, G.S, Brothers, M.C, Ho, M, Wilson, B.A, Rienstra, C.M. | | Deposit date: | 2015-12-12 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The membrane localization domains of two distinct bacterial toxins form a 4-helix-bundle in solution.
Protein Sci., 26, 2017
|
|
2N9V
 
 | | Solution NMR Structure of the membrane localization domain from Pasteurella multocida toxin | | Descriptor: | Dermonecrotic toxin | | Authors: | Hisao, G.S, Brothers, M.C, Ho, M, Wilson, B.A, Rienstra, C.M. | | Deposit date: | 2015-12-12 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The membrane localization domains of two distinct bacterial toxins form a 4-helix-bundle in solution.
Protein Sci., 26, 2017
|
|
4ECE
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with guanine | | Descriptor: | GUANINE, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-26 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
7CGY
 
 | | Human DMC1 Q244M mutant of the post-synaptic complexes | | Descriptor: | CALCIUM ION, Meiotic recombination protein DMC1/LIM15 homolog, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chi, H.Y, Ho, M.C, Tsai, M.D, Luo, S.C, Yeh, H.Y. | | Deposit date: | 2020-07-03 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C9C
 
 | | Human DMC1 pre-synaptic complexes | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Meiotic recombination protein DMC1/LIM15 homolog, ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7YPK
 
 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7C99
 
 | | Human DMC1 post-synaptic complexes with mismatched dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C9A
 
 | | Human RAD51 post-synaptic complexes mutant (V273P, D274G) | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Chi, H.Y, Ho, M.C, Tsai, M.D, Luo, S.C, Yeh, H.Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
7C98
 
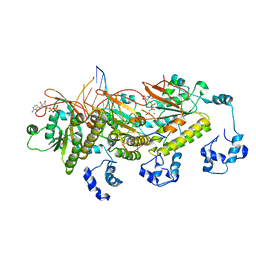 | | Human DMC1 post-synaptic complexes | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Luo, S.C, Yeh, H.Y, Chi, P, Ho, M.C, Tsai, M.D. | | Deposit date: | 2020-06-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures.
Nat Commun, 12, 2021
|
|
3D4P
 
 | |
7XDG
 
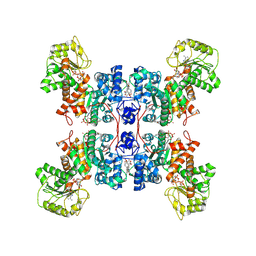 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor MDSA | | Descriptor: | 5-[(3-carboxy-4-oxidanyl-phenyl)methyl]-2-oxidanyl-benzoic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDF
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in a ternary complex with NAD+ and allosteric inhibitor EA | | Descriptor: | 4-[(3-carboxy-2-oxidanyl-naphthalen-1-yl)methyl]-3-oxidanyl-naphthalene-2-carboxylic acid, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
7XDE
 
 | | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form | | Descriptor: | NAD-dependent malic enzyme, mitochondrial, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Wang, C.H, Hsieh, J.T, Ho, M.C, Hung, H.C. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration
Commun Biol, 6, 2023
|
|
4GKA
 
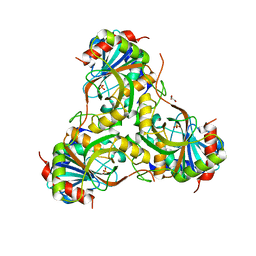 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with phosphate | | Descriptor: | GLYCEROL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
4EAR
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
3EWD
 
 | | Crystal structure of adenosine deaminase mutant (delta Asp172) from Plasmodium vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
5BYW
 
 | |
7YPI
 
 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPH
 
 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
3EWC
 
 | | Crystal Structure of adenosine deaminase from Plasmodial vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
5VOE
 
 | | DesGla-XaS195A Bound to Aptamer 11F7t | | Descriptor: | Aptamer 11F7t (36-MER), CALCIUM ION, Coagulation factor X, ... | | Authors: | Gunaratne, R, Kumar, S, Frederiksen, J.W, Stayrook, S, Lohrmann, J.L, Perry, K, Chabata, C.V, Thalji, N.K, Ho, M.D, Arepally, G, Camire, R.M, Krishnaswamy, S.K, Sullenger, B.A. | | Deposit date: | 2017-05-02 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of aptamer and drug for reversible anticoagulation in cardiopulmonary bypass.
Nat. Biotechnol., 36, 2018
|
|
2GO0
 
 | |
3WRE
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
