7SJ3
 
 | | Structure of CDK4-Cyclin D3 bound to abemaciclib | | Descriptor: | Cyclin-dependent kinase 4, G1/S-specific cyclin-D3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Hilgers, M.T, Pelletier, L.A. | | Deposit date: | 2021-10-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of CDK4-Cyclin D3 bound to abemaciclib
To Be Published
|
|
4QRE
 
 | |
1IE0
 
 | | CRYSTAL STRUCTURE OF LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, GLYCEROL, ZINC ION | | Authors: | Hilgers, M.T, Ludwig, M.L. | | Deposit date: | 2001-04-05 | | Release date: | 2001-10-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the quorum-sensing protein LuxS reveals a catalytic metal site.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4LAG
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 6-chloro-7-[5,6-dimethyl-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4LAH
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethyl-2-(1,3-thiazol-4-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4LAE
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethyl-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4LEK
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 7-[5,6-dimethoxy-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
4HWS
 
 | |
4HWP
 
 | |
4HWT
 
 | |
4HWO
 
 | |
4HWR
 
 | |
1Q85
 
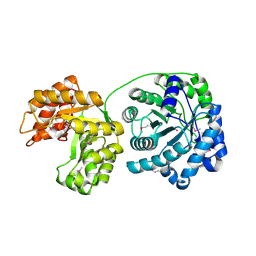 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex, Se-Met) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6U07
 
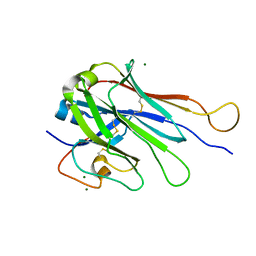 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
4QRD
 
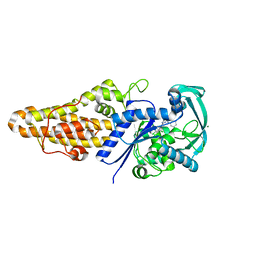 | | Structure of Methionyl-tRNA Synthetase in complex with N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Descriptor: | MAGNESIUM ION, Methionyl-tRNA synthetase, N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Authors: | Li, X, Hilgers, M.T, Stidham, M, Brown-Driver, V, Shaw, K.J, Finn, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and SAR of a Novel Series of Pyrimidine Antibacterials Targeting Methionyl-tRNA Synthetase
to be published
|
|
1P31
 
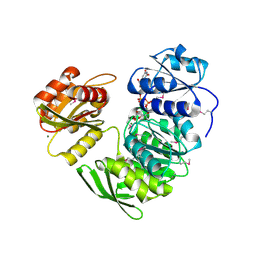 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1P3D
 
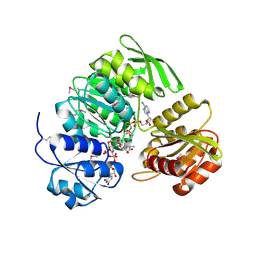 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
1Q8J
 
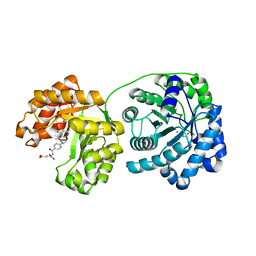 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+, Hcy, methyltetrahydrofolate complex) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, ... | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q7Q
 
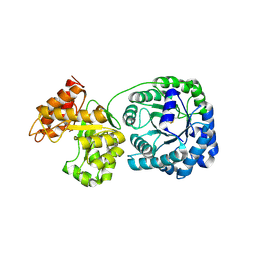 | | Cobalamin-dependent methionine synthase (1-566) from T. maritima (Oxidized, Orthorhombic) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q8A
 
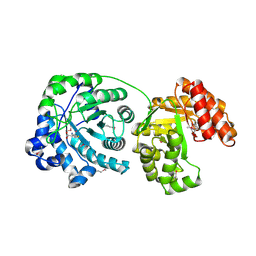 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+:L-Hcy complex, Se-Met) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q7M
 
 | | Cobalamin-dependent methionine synthase (MetH) from Thermotoga maritima (Oxidized, Monoclinic) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q7Z
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
