3I8X
 
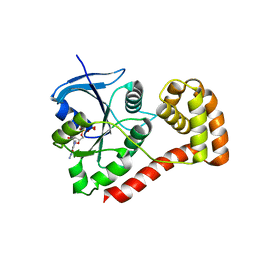 | | Structure of the cytosolic domain of E. coli FeoB, GDP-bound form | | Descriptor: | Ferrous iron transport protein B, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Petermann, N, Hansen, G, Hogg, T, Hilgenfeld, R. | | Deposit date: | 2009-07-10 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the intrinsic GTPase and GDI activities of FeoB, a prokaryotic transmembrane GTP/GDP-binding protein
To be Published
|
|
3IBY
 
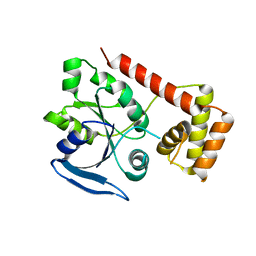 | |
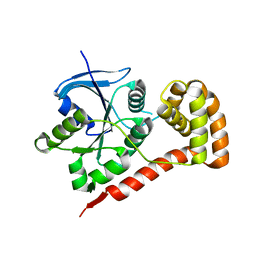
3I8S
 
 | |
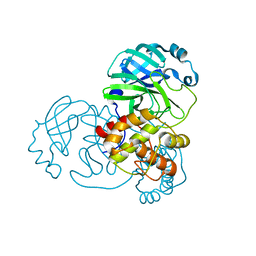
8C9L
 
 | |
8C9O
 
 | | Crystal structure of SARS-CoV-2 Mpro-S144A mutant in complex with 13b-K | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro_S144A mutant in complex with 13b-K
To Be Published
|
|
8C9P
 
 | |
8C9U
 
 | |
8C9Q
 
 | | Crystal structure of SARS-CoV-2 Mpro-Q189K mutant in complex with 13b-K | | Descriptor: | Non-structural protein 11, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro-Q189K mutant in complex with 13b-K
To Be Published
|
|
8CA8
 
 | |
8CA6
 
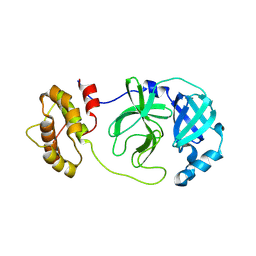 | |
8CAC
 
 | | Crystal structure of SARS-CoV-2 Mpro-H172Y mutant in complex with 13b-K | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro-H172Y mutant in complex with 13b-K
To Be Published
|
|
8CAJ
 
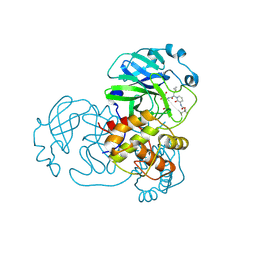 | | Crystal structure of SARS-CoV-2 Mpro-E166V mutant in complex with 13b-K | | Descriptor: | Non-structural protein 11, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{S})-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2023-01-24 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro-E166V mutant in complex with 13b-K
To Be Published
|
|
1F13
 
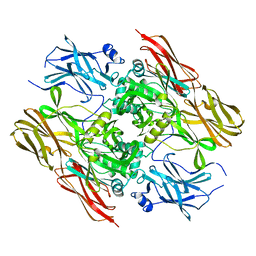 | |
2CIJ
 
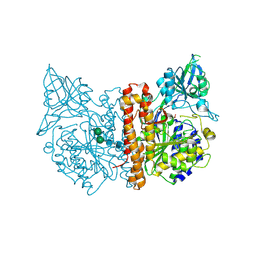 | | membrane-bound glutamate carboxypeptidase II (GCPII) with bound methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Barinka, C, Plechanovova, A, Rulisek, L, Mlcochova, P, Majer, P, Slusher, B.S, Hilgenfeld, R, Mesters, J.R, Konvalinka, J. | | Deposit date: | 2006-03-21 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutamate Carboxypeptidase II
Handbook of Metalloproteins, 4, 2011
|
|
1IZH
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
6Y2F
 
 | | Crystal structure (monoclinic form) of the complex resulting from the reaction between SARS-CoV-2 (2019-nCoV) main protease and tert-butyl (1-((S)-1-(((S)-4-(benzylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-3-cyclopropyl-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate (alpha-ketoamide 13b) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Sun, X, Hilgenfeld, R. | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
1IZI
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | Descriptor: | CHLORIDE ION, proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | Deposit date: | 2002-10-02 | | Release date: | 2002-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
6Y7M
 
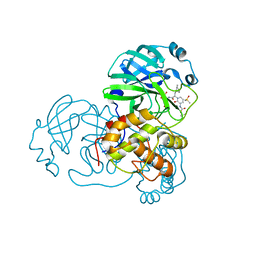 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Hilgenfeld, R. | | Deposit date: | 2020-03-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y2E
 
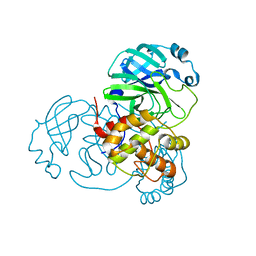 | |
2J98
 
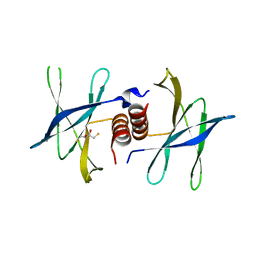 | | Human coronavirus 229E non structural protein 9 cys69ala mutant (Nsp9) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, REPLICASE POLYPROTEIN 1AB | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
2J97
 
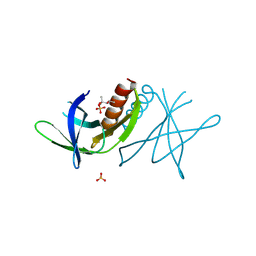 | | Human coronavirus 229E non structural protein 9 (Nsp9) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, REPLICASE POLYPROTEIN 1AB, SULFATE ION | | Authors: | Ponnusamy, R, Mesters, J.R, Moll, R, Hilgenfeld, R. | | Deposit date: | 2006-11-03 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Variable Oligomerization Modes in Coronavirus Non-Structural Protein 9.
J.Mol.Biol., 383, 2008
|
|
6Z2E
 
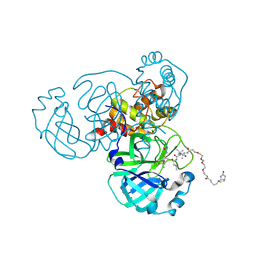 | | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone | | Descriptor: | (4~{S})-4-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[3-[2-[2-[2-[2-[5-[(3~{a}~{S},4~{R},6~{a}~{R})-2-oxidanylidene-3,3~{a},4,6~{a}-tetrahydro-1~{H}-thieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]butanoyl]amino]-3,3-dimethyl-butanoyl]amino]-4-methyl-pentanoyl]amino]-6-methylsulfonyl-hexanamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Zhang, L, Hilgenfeld, R. | | Deposit date: | 2020-05-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 Mpro in complex with the activity-based probe, biotin-PEG(4)-Abu-Tle-Leu-Gln-vinylsulfone
To Be Published
|
|
3EJG
 
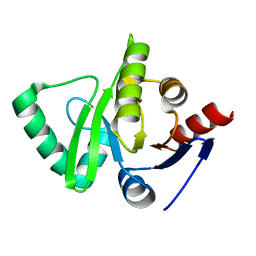 | | Crystal structure of HCoV-229E X-domain | | Descriptor: | Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-18 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3EKE
 
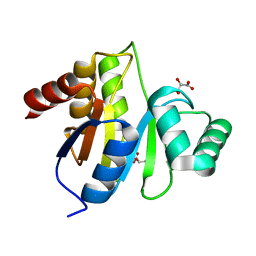 | | Crystal structure of IBV X-domain at pH 5.6 | | Descriptor: | L(+)-TARTARIC ACID, Non-structural protein 3 | | Authors: | Piotrowski, Y, Hansen, G, Hilgenfeld, R. | | Deposit date: | 2008-09-19 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the X-domains of a Group-1 and a Group-3 coronavirus reveal that ADP-ribose-binding may not be a conserved property.
Protein Sci., 18, 2009
|
|
3EJF
 
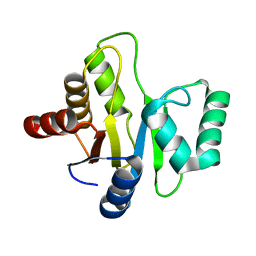 | |
