8CA6
 
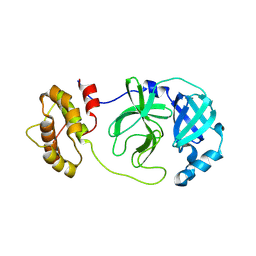 | |
8CAJ
 
 | | Crystal structure of SARS-CoV-2 Mpro-E166V mutant in complex with 13b-K | | 分子名称: | Non-structural protein 11, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{S})-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | 著者 | El Kilani, H, Hilgenfeld, R. | | 登録日 | 2023-01-24 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 Mpro-E166V mutant in complex with 13b-K
To Be Published
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | 分子名称: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
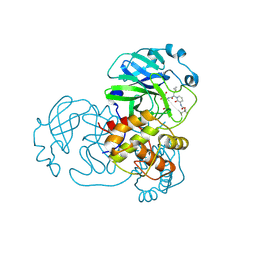
1LK9
 
 | | The Three-dimensional Structure of Alliinase from Garlic | | 分子名称: | 2-AMINO-ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Kuettner, E.B, Hilgenfeld, R, Weiss, M.S. | | 登録日 | 2002-04-24 | | 公開日 | 2002-12-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | The active principle of garlic at atomic resolution
J.Biol.Chem., 277, 2002
|
|
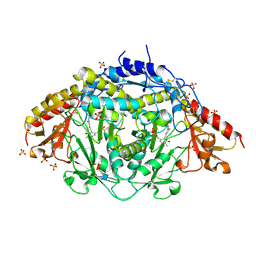
1EXM
 
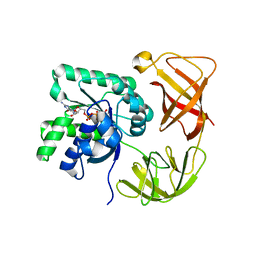 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | 分子名称: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | 登録日 | 2000-05-03 | | 公開日 | 2000-06-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
4QL6
 
 | |
2JBK
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with quisqualic acid (quisqualate, alpha-amino-3,5-dioxo-1,2,4- oxadiazolidine-2-propanoic acid) | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mesters, J.R, Henning, K, Hilgenfeld, R. | | 登録日 | 2006-12-07 | | 公開日 | 2006-12-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2JBJ
 
 | | membrane-bound glutamate carboxypeptidase II (GCPII) in complex with 2-PMPA (2-phosphonoMethyl-pentanedioic acid) | | 分子名称: | (2S)-2-(PHOSPHONOMETHYL)PENTANEDIOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Mesters, J.R, Henning, K, Hilgenfeld, R. | | 登録日 | 2006-12-07 | | 公開日 | 2006-12-18 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Human Glutamate Carboxypeptidase II Inhibition: Structures of Gcpii in Complex with Two Potent Inhibitors, Quisqualate and 2-Pmpa.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4K35
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | 著者 | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | 登録日 | 2013-04-10 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | 分子名称: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | 著者 | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | 登録日 | 2013-04-10 | | 公開日 | 2013-10-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5HIH
 
 | |
2XCU
 
 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP | | 分子名称: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | 登録日 | 2010-04-26 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HOL
 
 | |
2XCI
 
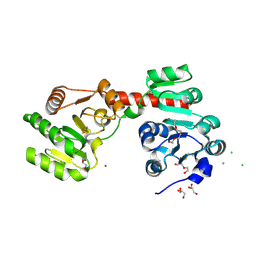 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | 分子名称: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | 著者 | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | 登録日 | 2010-04-26 | | 公開日 | 2011-05-11 | | 最終更新日 | 2019-05-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5N19
 
 | |
5NGN
 
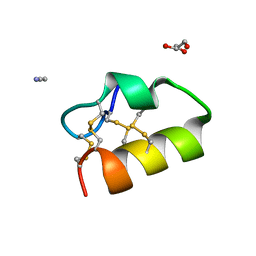 | | Lybatide 2, a cystine-rich peptide from Lycium barbarum | | 分子名称: | ACETONITRILE, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | 著者 | Lei, J, Tan, W.L, Sakai, N, Hilgenfeld, R. | | 登録日 | 2017-03-18 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Lybatides from Lycium barbarum Contain An Unusual Cystine-stapled Helical Peptide Scaffold.
Sci Rep, 7, 2017
|
|
5NFS
 
 | |
3I92
 
 | | Structure of the cytosolic domain of E. coli FeoB, GppCH2p-bound form | | 分子名称: | Ferrous iron transport protein B, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | 著者 | Petermann, N, Hansen, G, Hogg, T, Hilgenfeld, R. | | 登録日 | 2009-07-10 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the intrinsic GTPase and GDI activities of FeoB, a prokaryotic transmembrane GTP/GDP-binding protein
To be Published
|
|
3I8X
 
 | | Structure of the cytosolic domain of E. coli FeoB, GDP-bound form | | 分子名称: | Ferrous iron transport protein B, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Petermann, N, Hansen, G, Hogg, T, Hilgenfeld, R. | | 登録日 | 2009-07-10 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural basis for the intrinsic GTPase and GDI activities of FeoB, a prokaryotic transmembrane GTP/GDP-binding protein
To be Published
|
|
3IBY
 
 | |
3I8S
 
 | |
5NH0
 
 | |
1F13
 
 | |
5N5O
 
 | |
1IZH
 
 | | Inhibitor of HIV protease with unusual binding mode potently inhibiting multi-resistant protease mutants | | 分子名称: | proteinase, {(1S)-1-BENZYL-4-[3-CARBAMOYL-1-(1-CARBAMOYL-2-PHENYL-ETHYLCARBAMOYL)-(S)-PROPYLCARBAMOYL]-2-OXO-5-PHENYL-PENTYL}-CARBAMIC ACID TERT-BUTYL ESTER | | 著者 | Weber, J, Mesters, J.R, Lepsik, M, Prejdova, J, Svec, M, Sponarova, J, Mlcochova, P, Skalicka, K, Strisovsky, K, Uhlikova, T, Soucek, M, Machala, L, Stankova, M, Vondrasek, J, Klimkait, T, Kraeusslich, H.-G, Hilgenfeld, R, Konvalinka, J. | | 登録日 | 2002-10-02 | | 公開日 | 2002-12-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unusual Binding Mode of an HIV-1 Protease Inhibitor Explains its Potency against Multi-drug-resistant Virus Strains
J.MOL.BIOL., 324, 2002
|
|
