2ZZT
 
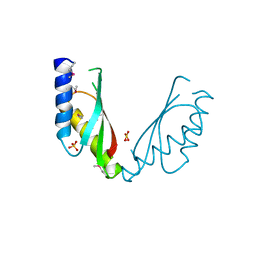 | | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Higuchi, T, Hattori, M, Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2009-02-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | Crystal structure of the cytosolic domain of the cation diffusion facilitator family protein
Proteins, 76, 2009
|
|
3ALW
 
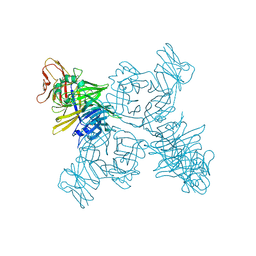 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I, MV-H-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALZ
 
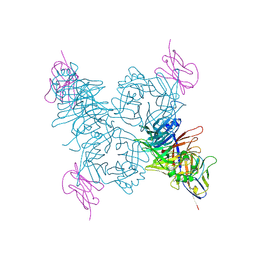 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (Form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CDw150, Hemagglutinin | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.515 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
3ALX
 
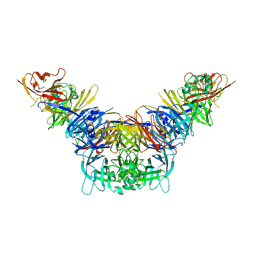 | | Crystal structure of the measles virus hemagglutinin bound to its cellular receptor SLAM (MV-H(L482R)-SLAM(N102H/R108Y) fusion) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin,LINKER,CDw150 | | Authors: | Hashiguchi, T, Ose, T, Kubota, M, Maita, N, Kamishikiryo, J, Maenaka, K, Yanagi, Y. | | Deposit date: | 2010-08-09 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM
Nat.Struct.Mol.Biol., 18, 2011
|
|
5UQY
 
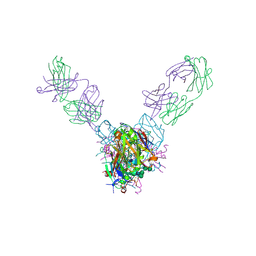 | | Crystal structure of Marburg virus GP in complex with the human survivor antibody MR78 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP1, ... | | Authors: | Hashiguchi, T, Fusco, M.L, Hastie, K.M, Bomholdt, Z.A, Lee, J.E, Flyak, A.I, Matsuoka, R, Kohda, D, Yanagi, Y, Hammel, M, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160, 2015
|
|
5YXW
 
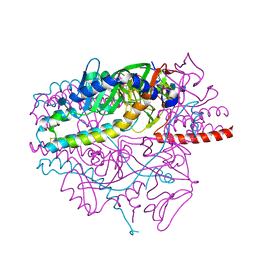 | | Crystal structure of the prefusion form of measles virus fusion protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-07 | | Release date: | 2018-02-21 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZD
 
 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor peptide (FIP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.636 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YZC
 
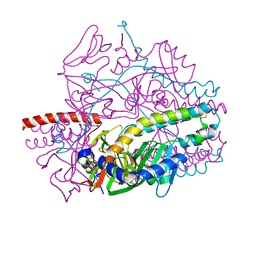 | | Crystal structure of the prefusion form of measles virus fusion protein in complex with a fusion inhibitor compound (AS-48) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitro-2-[(phenylacetyl)amino]benzamide, ... | | Authors: | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2ZB6
 
 | | Crystal structure of the measles virus hemagglutinin (oligo-sugar type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ZB5
 
 | | Crystal structure of the measles virus hemagglutinin (complex-sugar-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8HGM
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, NIV-11 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
8HES
 
 | | Crystal structure of SARS-CoV-2 RBD and NIV-10 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-10 Fab H-chain, NIV-10 Fab L-chain, ... | | Authors: | Moriyama, S, Anraku, Y, Taminishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5X60
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 9 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2017-02-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
5H6Q
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
5H6R
 
 | | Crystal structure of LSD1-CoREST in complex with peptide 13 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kkuchi, M, Amano, Y, Sato, S, Yokoyama, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2016-11-14 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Development and crystallographic evaluation of histone H3 peptide with N-terminal serine substitution as a potent inhibitor of lysine-specific demethylase 1.
Bioorg. Med. Chem., 25, 2017
|
|
3ABU
 
 | | Crystal Structure of LSD1 in complex with a 2-PCPA derivative, S1201 | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3R,3aS)-3-[2-(benzyloxy)-3-fluorophenyl]-1-hydroxy-10,11-dimethyl-4,6-dioxo-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
3ABT
 
 | | Crystal Structure of LSD1 in complex with trans-2-pentafluorophenylcyclopropylamine | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(1R,3R,3aS)-1-hydroxy-10,11-dimethyl-4,6-dioxo-3-(pentafluorophenyl)-2,3,5,6-tetrahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]pentyl dihydrogen diphosphate | | Authors: | Mimasu, S, Umezawa, N, Sato, S, Higuchi, T, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structurally Designed trans-2-Phenylcyclopropylamine Derivatives Potently Inhibit Histone Demethylase LSD1/KDM1
Biochemistry, 49, 2010
|
|
1ART
 
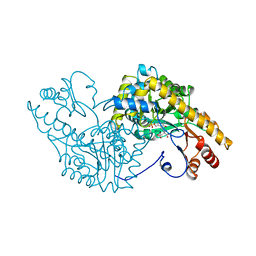 | |
1ARS
 
 | |
7CDC
 
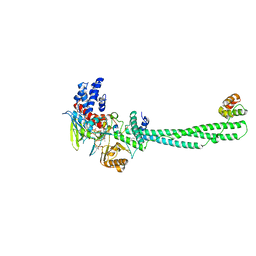 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRP peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDF
 
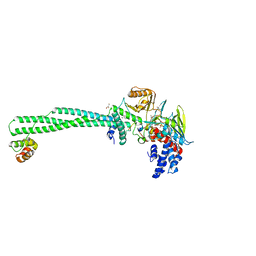 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRK peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDD
 
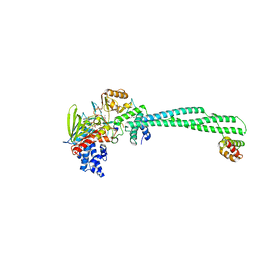 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
