3CA8
 
 | | Crystal structure of Escherichia coli YdcF, an S-adenosyl-L-methionine utilizing enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Protein ydcF, SULFATE ION | | Authors: | Lim, K, Chao, K, Lehmann, C, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2008-02-19 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Escherichia coli YdcF binds S-adenosyl-L-methionine and adopts an alpha/beta-fold characteristic of nucleotide-utilizing enzymes.
Proteins, 72, 2008
|
|
3FA4
 
 | | Crystal structure of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member, triclinic crystal form | | Descriptor: | 2,3-dimethylmalate lyase, MAGNESIUM ION | | Authors: | Narayanan, B.C, Herzberg, O. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure and function of 2,3-dimethylmalate lyase, a PEP mutase/isocitrate lyase superfamily member.
J.Mol.Biol., 386, 2009
|
|
1PIO
 
 | |
1PCH
 
 | |
1GGO
 
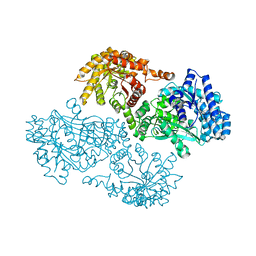 | | T453A MUTANT OF PYRUVATE, PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE, PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 2000-08-29 | | Release date: | 2001-01-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of domain-domain docking sites within Clostridium symbiosum pyruvate phosphate dikinase by amino acid replacement.
J.Biol.Chem., 275, 2000
|
|
4OJ6
 
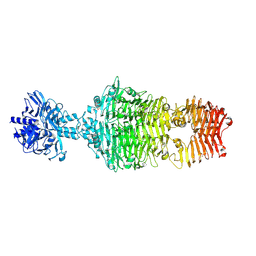 | |
4OJO
 
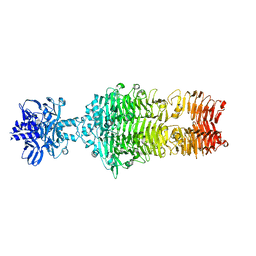 | |
4OJP
 
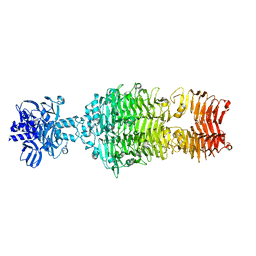 | |
2HJP
 
 | |
2ISW
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
2ISV
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Galkin, A, Herzberg, O. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization, kinetics, and crystal structures of fructose-1,6-bisphosphate aldolase from the human parasite, Giardia lamblia.
J.Biol.Chem., 282, 2007
|
|
4OJ5
 
 | |
4OJL
 
 | |
2GPR
 
 | | GLUCOSE PERMEASE IIA FROM MYCOPLASMA CAPRICOLUM | | Descriptor: | GLUCOSE-PERMEASE IIA COMPONENT | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A promiscuous binding surface: crystal structure of the IIA domain of the glucose-specific permease from Mycoplasma capricolum.
Structure, 6, 1998
|
|
2HWG
 
 | | Structure of phosphorylated Enzyme I of the phosphoenolpyruvate:sugar phosphotransferase system | | Descriptor: | MAGNESIUM ION, OXALATE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Lim, K, Teplyakov, A, Herzberg, O. | | Deposit date: | 2006-08-01 | | Release date: | 2006-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of phosphorylated enzyme I, the phosphoenolpyruvate:sugar phosphotransferase system sugar translocation signal protein.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HRW
 
 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
1ZNB
 
 | | METALLO-BETA-LACTAMASE | | Descriptor: | METALLO-BETA-LACTAMASE, SODIUM ION, ZINC ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis.
Structure, 4, 1996
|
|
1X6J
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1X6I
 
 | | Crystal structure of ygfY from Escherichia coli | | Descriptor: | Hypothetical protein ygfY | | Authors: | Lim, K, Doseeva, V, Sarikaya Demirkan, E, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-11 | | Release date: | 2005-02-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the YgfY from Escherichia coli, a protein that may be involved in transcriptional regulation
Proteins, 58, 2005
|
|
1VDR
 
 | | DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, PHOSPHATE ION | | Authors: | Pieper, U, Herzberg, O. | | Deposit date: | 1997-11-30 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii.
Structure, 6, 1998
|
|
1XG4
 
 | | Crystal Structure of the C123S 2-Methylisocitrate Lyase Mutant from Escherichia coli in complex with the inhibitor isocitrate | | Descriptor: | ISOCITRIC ACID, MAGNESIUM ION, Probable methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
1XG3
 
 | | Crystal structure of the C123S 2-methylisocitrate lyase mutant from Escherichia coli in complex with the reaction product, Mg(II)-pyruvate and succinate | | Descriptor: | MAGNESIUM ION, PYRUVIC ACID, Probable methylisocitrate lyase, ... | | Authors: | Liu, S, Lu, Z, Han, Y, Melamud, E, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of 2-Methylisocitrate Lyase in Complex with Product and with Isocitrate Inhibitor Provide Insight into Lyase Substrate Specificity, Catalysis and Evolution
Biochemistry, 44, 2005
|
|
1ZLP
 
 | | Petal death protein PSR132 with cysteine-linked glutaraldehyde forming a thiohemiacetal adduct | | Descriptor: | 5-HYDROXYPENTANAL, MAGNESIUM ION, petal death protein | | Authors: | Teplyakov, A, Liu, S, Lu, Z, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-05-08 | | Release date: | 2006-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Petal Death Protein from Carnation Flower.
Biochemistry, 44, 2005
|
|
2AAF
 
 | |
1JDE
 
 | | K22A mutant of pyruvate, phosphate dikinase | | Descriptor: | PYRUVATE, PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Ye, D, Wei, M, McGuire, M, Huang, K, Kapadia, G, Herzberg, O, Martin, B.M, Dunaway-Mariano, D. | | Deposit date: | 2001-06-13 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of the catalytic site within the ATP-grasp domain of Clostridium symbiosum pyruvate phosphate dikinase.
J.Biol.Chem., 276, 2001
|
|
