2ABR
 
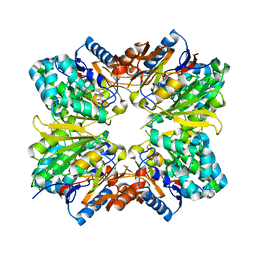 | |
4V7N
 
 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
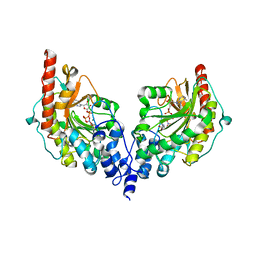
2DIK
 
 | | R337A MUTANT OF PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PROTEIN (PYRUVATE PHOSPHATE DIKINASE), SULFATE ION | | Authors: | Huang, K, Herzberg, O. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Location of the phosphate binding site within Clostridium symbiosum pyruvate phosphate dikinase.
Biochemistry, 37, 1998
|
|
3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
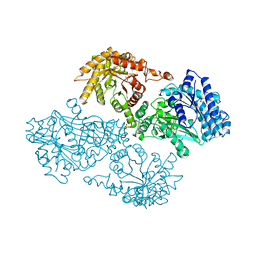
4JZ7
 
 | | Carbamate kinase from Giardia lamblia bound to AMP-PNP | | Descriptor: | Carbamate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lim, K, Herzberg, O. | | Deposit date: | 2013-04-02 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Carbamate Kinase from Giardia lamblia Bound with Citric Acid and AMP-PNP.
Plos One, 8, 2013
|
|
7PWO
 
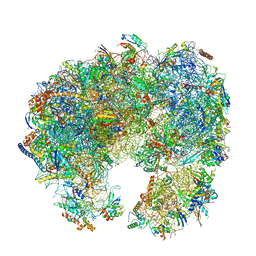 | | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S26, 40S ribosomal protein S30, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7PWG
 
 | | Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L18a, 60S ribosomal protein L27, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
4JZ9
 
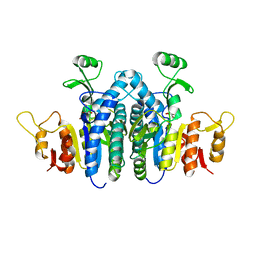 | |
4JZ8
 
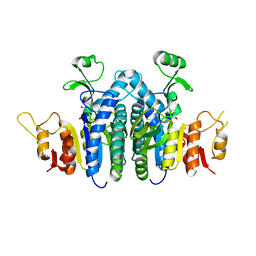 | |
7PWF
 
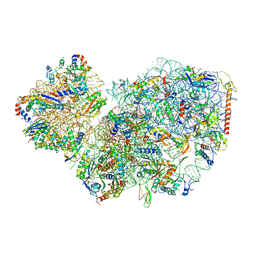 | | Cryo-EM structure of small subunit of Giardia lamblia ribosome at 2.9 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S25, 40S ribosomal protein S26, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
8P33
 
 | | BB0238 from Borrelia burgdorferi | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
8P32
 
 | | BB0238 from Borrelia burgdorferi, Se-Met data for Leu240Met mutant | | Descriptor: | BB0238 | | Authors: | Brangulis, K, Foor, S.D, Shakya, A.K, Rana, V.S, Bista, S, Kitsou, C, Ronzetti, M, Linden, S.B, Altieri, A.S, Akopjana, I, Baljinnyam, B, Nelson, D.C, Simeonov, A, Herzberg, O, Caimano, M.J, Pal, U. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A unique borrelial protein facilitates microbial immune evasion.
Mbio, 14, 2023
|
|
7REJ
 
 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-335, obtained in the presence of NaK-Tartrate | | Descriptor: | IMIDAZOLE, Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-13 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
4FWW
 
 | |
2ZNB
 
 | | METALLO-BETA-LACTAMASE (CADMIUM-BOUND FORM) | | Descriptor: | CADMIUM ION, METALLO-BETA-LACTAMASE, SODIUM ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1997-10-14 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the cadmium- and mercury-substituted metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 6, 1997
|
|
1ALQ
 
 | |
2PE4
 
 | | Structure of Human Hyaluronidase 1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Chao, K.L, Herzberg, O. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Human Hyaluronidase-1, a Hyaluronan Hydrolyzing Enzyme Involved in Tumor Growth and Angiogenesis
Biochemistry, 46, 2007
|
|
7RFO
 
 | | SeMet Tailspike protein 4 (TSP4) phage CBA120, residues 1-335, obtained in the presence of LiSO4 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
7RFV
 
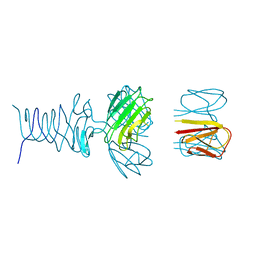 | | Tailspike protein 4 (TSP4) from phage CBA120, residues 1-250, obtained in the presence of PEG8000 | | Descriptor: | Tailspike protein | | Authors: | Chao, K, Shang, X, Grenfield, J, Linden, S.B, Nelson, D.C, Herzberg, O. | | Deposit date: | 2021-07-14 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Escherichia coli O157:H7 bacteriophage CBA120 tailspike protein 4 baseplate anchor and tailspike assembly domains (TSP4-N).
Sci Rep, 12, 2022
|
|
4GAH
 
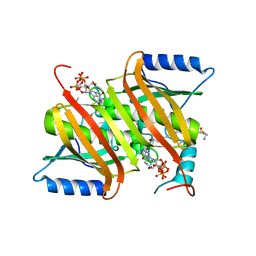 | | Human acyl-CoA thioesterases 4 in complex with undecan-2-one-CoA inhibitor | | Descriptor: | Thioesterase superfamily member 4, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-[2-[(2R)-2-oxidanylundecyl]sulfanylethylamino]propyl]amino]butyl] hydrogen phosphate | | Authors: | Lim, K, Pathak, M.C, Herzberg, O. | | Deposit date: | 2012-07-25 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation of structure and function in the human hotdog-fold enzyme hTHEM4.
Biochemistry, 51, 2012
|
|
2OUT
 
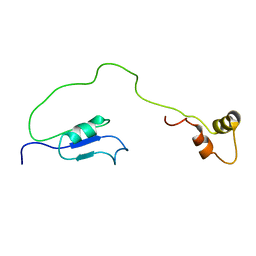 | | Solution Structure of HI1506, a Novel Two Domain Protein from Haemophilus influenzae | | Descriptor: | Mu-like prophage FluMu protein gp35, Protein HI1507 in Mu-like prophage FluMu region | | Authors: | Sari, N, He, Y, Doseeva, V, Surabian, K, Schwarz, F, Herzberg, O, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2007-02-12 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HI1506, a novel two-domain protein from Haemophilus influenzae.
Protein Sci., 16, 2007
|
|
4OLC
 
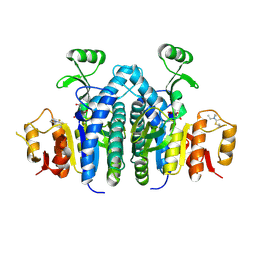 | |
1DJC
 
 | |
3QYN
 
 | |
1DJA
 
 | |
