3P9T
 
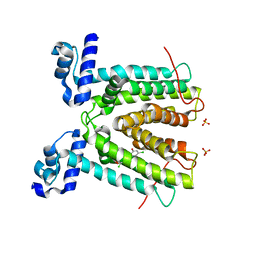 | | SmeT-Triclosan complex | | Descriptor: | Repressor, SULFATE ION, TRICLOSAN | | Authors: | Hernandez, A, Ruiz, F.M, Romero, A, Martinez, J.L. | | Deposit date: | 2010-10-18 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Binding of Triclosan to SmeT, the Repressor of the Multidrug Efflux Pump SmeDEF, Induces Antibiotic Resistance in Stenotrophomonas maltophilia.
Plos Pathog., 7, 2011
|
|
5C7W
 
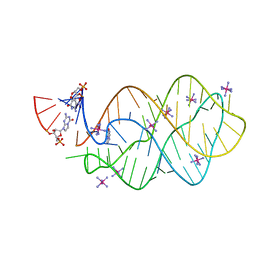 | | 5'-monophosphate Z:P Guanine Riboswitch bound to hypoxanthine. | | Descriptor: | 5'-monophosphate Z:P guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | Authors: | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5C7U
 
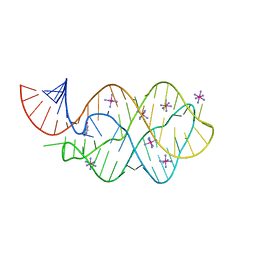 | | 5'-monophosphate wt Guanine Riboswitch bound to hypoxanthine. | | Descriptor: | 5'-monophosphate wt guanine riboswitch, COBALT HEXAMMINE(III), HYPOXANTHINE | | Authors: | Hernandez, A.R, Shao, Y, Hoshika, S, Yang, Z, Shelke, S.A, Herrou, J, Kim, H.-J, Kim, M.-J, Piccirilli, J.A, Benner, S.A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A Crystal Structure of a Functional RNA Molecule Containing an Artificial Nucleobase Pair.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6USV
 
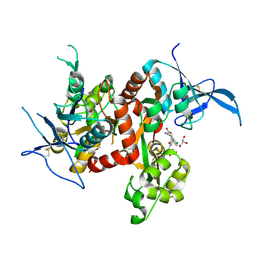 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
7KOT
 
 | |
7KP5
 
 | |
6TJV
 
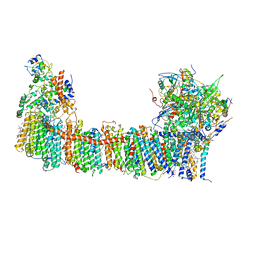 | | Structure of the NDH-1MS complex from Thermosynechococcus elongatus | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, BETA-CAROTENE, ... | | Authors: | Schuller, J.M, Saura, P, Thiemann, J, Schuller, S.K, Gamiz-Hernandez, A.P, Kurisu, G, Nowaczyk, M.M, Kaila, V.R.I. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Redox-coupled proton pumping drives carbon concentration in the photosynthetic complex I.
Nat Commun, 11, 2020
|
|
7MXL
 
 | |
8TG9
 
 | |
8OVC
 
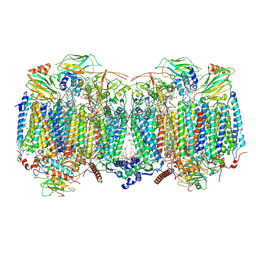 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
8OVD
 
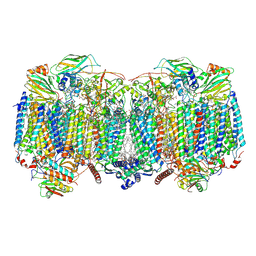 | | Respiratory supercomplex (III2-IV2) from Mycobacterium smegmatis | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Krol, S, Sjostrand, D, Riepl, D, Gamiz-Hernandez, A, Brzezinski, P, Kaila, V, Hogbom, M. | | Deposit date: | 2023-04-25 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Long-range charge transfer mechanism of the III 2 IV 2 mycobacterial supercomplex.
Nat Commun, 15, 2024
|
|
8TGA
 
 | |
8A6T
 
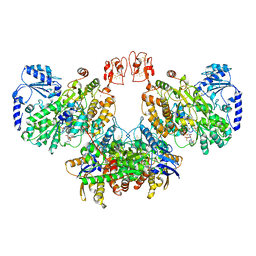 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, Electron bifurcating hydrogenase subunit HydA1, Electron bifurcating hydrogenase subunit HydB, ... | | Authors: | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-06-19 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8A5E
 
 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Acetobacterium woodii in the reduced state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8EYM
 
 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM SHEWANELLA DENITRIFICANS OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AND N-ACETYLGLUCOSAMINE-6-PHOSPHATE AT 2.31 A RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rodriguez-Hernandez, A, Marcos-Viquez, J, Rodriguez-Romero, A, Bustos-Jaimes, I. | | Deposit date: | 2022-10-27 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
8EOL
 
 | |
8FDB
 
 | | CRYSTAL STRUCTURE OF NAGB-II PHOSPHOSUGAR ISOMERASE FROM Shewanella denitrificans OS217 IN COMPLEX WITH GLUCITOLAMINE-6-PHOSPHATE AT 3.06 A RESOLUTION. | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLYCEROL, Glutamine-fructose-6-phosphate transaminase (Isomerizing), ... | | Authors: | Rodriguez-Romero, A, Rodriguez-Hernandez, A, Marcos-Viquez, J, Bustos-Jaimes, I. | | Deposit date: | 2022-12-02 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Substrate binding in the allosteric site mimics homotropic cooperativity in the SIS-fold glucosamine-6-phosphate deaminases.
Protein Sci., 32, 2023
|
|
7R6Y
 
 | |
4JXZ
 
 | |
7M42
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of neutralizing antibodies REGN10985 and REGN10989 | | Descriptor: | REGN10985 antibody Fab fragment heavy chain, REGN10985 antibody Fab fragment light chain, REGN10989 antibody Fab fragment heavy chain, ... | | Authors: | Zhou, Y, Romero Hernandez, A, Saotome, K, Franklin, M.C. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies.
Cell, 184, 2021
|
|
4JXX
 
 | |
4JYZ
 
 | |
7N0V
 
 | |
6Z0C
 
 | | Structure of in silico modelled artificial Maquette-3 protein | | Descriptor: | Maquette-3, POTASSIUM ION | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M.E, Asami, S, Mader, S.L, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of buried charged networks in artificial proteins
Nat Commun, 12, 2021
|
|
