3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | 分子名称: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | 著者 | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | 登録日 | 2011-04-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1ORI
 
 | |
5SZX
 
 | | Epstein-Barr virus Zta DNA binding domain homodimer in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*GP*(5CM)P*GP*AP*TP*GP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*AP*TP*(5CM)P*GP*CP*TP*CP*AP*GP*TP*GP*CP*T)-3'), PHOSPHATE ION, ... | | 著者 | Hong, S, Horton, J.R, Cheng, X. | | 登録日 | 2016-08-15 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.
Nucleic Acids Res., 45, 2017
|
|
1OR8
 
 | |
5T0U
 
 | | CTCF ZnF2-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*CP*CP*TP*CP*AP*CP*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*AP*GP*TP*GP*AP*GP*G)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-08-16 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.199 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
4YVP
 
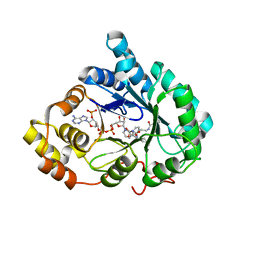 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | 分子名称: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
5DZM
 
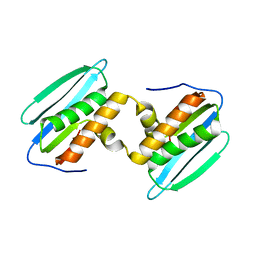 | |
1Q0T
 
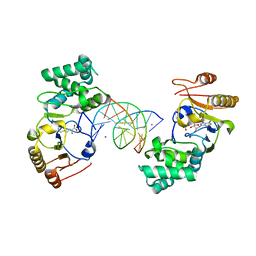 | | Ternary Structure of T4DAM with AdoHcy and DNA | | 分子名称: | 5'-D(*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*T)-3', DNA adenine methylase, IODIDE ION, ... | | 著者 | Yang, Z, Horton, J.R, Zhou, L, Zhang, X.J, Dong, A, Zhang, X, Schlagman, S.L, Kossykh, V, Hattman, S, Cheng, X. | | 登録日 | 2003-07-17 | | 公開日 | 2003-09-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of the bacteriophage T4 DNA adenine methyltransferase
Nat.Struct.Biol., 10, 2003
|
|
3RC0
 
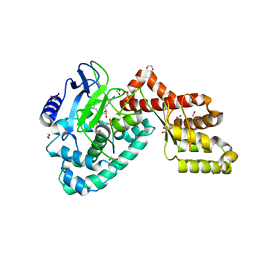 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
1F3L
 
 | |
1MR3
 
 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | 分子名称: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | 著者 | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | 登録日 | 2002-09-17 | | 公開日 | 2002-11-20 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
1MVX
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | 分子名称: | CRYPTIC LOCI REGULATOR 4, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | 登録日 | 2002-09-26 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
4EW0
 
 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
1MVH
 
 | | structure of the SET domain histone lysine methyltransferase Clr4 | | 分子名称: | Cryptic loci regulator 4, NICKEL (II) ION, SULFATE ION, ... | | 著者 | Min, J.R, Zhang, X, Cheng, X.D, Grewal, S.I.S, Xu, R.-M. | | 登録日 | 2002-09-25 | | 公開日 | 2002-10-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the SET domain histone lysine methyltransferase Clr4.
Nat.Struct.Biol., 9, 2002
|
|
5B73
 
 | |
4EVV
 
 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
3PTR
 
 | | PHF2 Jumonji domain | | 分子名称: | 1,2-ETHANEDIOL, PHD finger protein 2 | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.954 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
3PUA
 
 | | PHF2 Jumonji-NOG-Ni(II) | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | 著者 | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2010-12-03 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
4EW4
 
 | | mouse MBD4 glycosylase domain in complex with DNA containing a ribose sugar | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*GP*(3DR)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), Methyl-CpG-binding domain protein 4, ... | | 著者 | Hashimoto, H, Zhang, X, Cheng, X. | | 登録日 | 2012-04-26 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.791 Å) | | 主引用文献 | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4RNX
 
 | | K154 Circular Permutation of Old Yellow Enzyme | | 分子名称: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1 | | 著者 | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | 登録日 | 2014-10-26 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4N9C
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | 著者 | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | 登録日 | 2014-10-26 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.677 Å) | | 主引用文献 | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
4RNV
 
 | | G303 Circular Permutation of Old Yellow Enzyme with the Inhibitor p-Hydroxybenzaldehyde | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, P-HYDROXYBENZALDEHYDE | | 著者 | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | 登録日 | 2014-10-26 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.473 Å) | | 主引用文献 | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
2CSN
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH CKI7 | | 分子名称: | CASEIN KINASE-1, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, SULFATE ION | | 著者 | Xu, R.-M, Cheng, X. | | 登録日 | 1995-10-11 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for selectivity of the isoquinoline sulfonamide family of protein kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
