2HD7
 
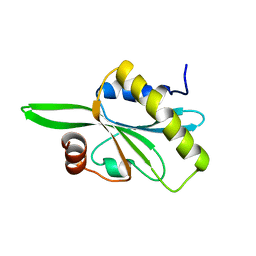 | | Solution structure of C-teminal domain of twinfilin-1. | | Descriptor: | Twinfilin-1 | | Authors: | Hellman, M.H, Paavilainen, V.O, Annila, A, Lappalainen, P, Permi, P.I. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis and evolutionary origin of actin filament capping by twinfilin
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1WM4
 
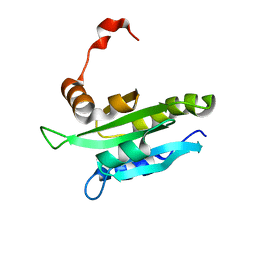 | | Solution structure of mouse coactosin, an actin filament binding protein | | Descriptor: | Coactosin-like protein | | Authors: | Hellman, M, Paavilainen, V.O, Naumanen, P, Lappalainen, P, Annila, A, Permi, P. | | Deposit date: | 2004-07-03 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins
Febs Lett., 576, 2004
|
|
2KVD
 
 | |
2KVE
 
 | |
5I22
 
 | | Amphiphysin SH3 in complex with Chikungunya virus nsP3 peptide | | Descriptor: | CHIKV nsP3 peptide, Myc box-dependent-interacting protein 1 | | Authors: | Tossavainen, H, Aitio, O, Hellman, M, Saksela, K, Permi, P. | | Deposit date: | 2016-02-04 | | Release date: | 2016-06-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the High Affinity Interaction between the Alphavirus Nonstructural Protein-3 (nsP3) and the SH3 Domain of Amphiphysin-2.
J.Biol.Chem., 291, 2016
|
|
5N9Q
 
 | | Structure of A. thaliana RCD1(468-567) | | Descriptor: | Inactive poly [ADP-ribose] polymerase RCD1 | | Authors: | Tossavainen, H, Hellman, M, Vainonen, J, Kangasjarvi, J, Permi, P. | | Deposit date: | 2017-02-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arabidopsis RCD1 coordinates chloroplast and mitochondrial functions through interaction with ANAC transcription factors.
Elife, 8, 2019
|
|
2ROL
 
 | | Structural Basis of PxxDY motif recognition in SH3 binding | | Descriptor: | 12-meric peptide from T-cell surface glycoprotein CD3 epsilon chain, Epidermal growth factor receptor kinase substrate 8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Paakkonen, K, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding
J.Mol.Biol., 382, 2008
|
|
2KXC
 
 | | 1H, 13C, and 15N Chemical Shift Assignments for IRTKS-SH3 and EspFu-R47 complex | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, EspF-like protein | | Authors: | Aitio, O, Hellman, M, Kazlauskas, A, Vingadassalom, D.F, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2010-04-30 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of tandem PxxP motifs as a unique Src homology 3-binding mode triggers pathogen-driven actin assembly
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2LNH
 
 | | Enterohaemorrhagic E. coli (EHEC) exploits a tryptophan switch to hijack host F-actin assembly | | Descriptor: | Brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1, Neural Wiskott-Aldrich syndrome protein, Secreted effector protein EspF(U) | | Authors: | Aitio, O, Hellman, M, Skehan, B, Kesti, T, Leong, J.M, Saksela, K, Permi, P. | | Deposit date: | 2011-12-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enterohaemorrhagic Escherichia coli exploits a tryptophan switch to hijack host f-actin assembly.
Structure, 20, 2012
|
|
2K2M
 
 | | Structural Basis of PxxDY Motif Recognition in SH3 Binding | | Descriptor: | Eps8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding.
J.Mol.Biol., 382, 2008
|
|
