1UWP
 
 | |
6MEL
 
 | | Succinyl-CoA synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6CY6
 
 | | Crystal structure of spermidine/spermine N-acetyltransferase SpeG from Escherichia coli in complex with tris(hydroxymethyl)aminomethane. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-04 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6D0G
 
 | | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | BROMIDE ION, MANGANESE (II) ION, Pirin family protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 1.78 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii.
To be Published
|
|
3HK1
 
 | | Identification and Characterization of a Small Molecule Inhibitor of Fatty Acid Binding Proteins | | Descriptor: | 4-{[2-(methoxycarbonyl)-5-(2-thienyl)-3-thienyl]amino}-4-oxo-2-butenoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Hertzel, A.V, Hellberg, K, Reynolds, J.M, Kruse, A.C, Juhlmann, B.E, Smith, A.J, Sanders, M.A, Ohlendorf, D.H, Suttles, J, Bernlohr, D.A. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and characterization of a small molecule inhibitor of Fatty Acid binding proteins.
J.Med.Chem., 52, 2009
|
|
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
1KOU
 
 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6D7Y
 
 | | 1.75 Angstrom Resolution Crystal Structure of the Toxic C-Terminal Tip of CdiA from Pseudomonas aeruginosa in Complex with Immune Protein | | Descriptor: | Hemagglutinin, immune protein | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Allen, J.P, Hauser, A.R, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-25 | | Release date: | 2019-05-01 | | Last modified: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A comparative genomics approach identifies contact-dependent growth inhibition as a virulence determinant.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6E1X
 
 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
6DT4
 
 | | 1.8 Angstrom Resolution Crystal Structure of cAMP-Regulatory Protein from Yersinia pestis in Complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, Cyclic AMP receptor protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Ritzert, J.T.H, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Cyclic AMP Receptor Protein Regulates Quorum Sensing and Global Gene Expression in Yersinia pestis during Planktonic Growth and Growth in Biofilms.
Mbio, 10, 2019
|
|
6DFU
 
 | | Tryptophan--tRNA ligase from Haemophilus influenzae. | | Descriptor: | TRYPTOPHAN, Tryptophan--tRNA ligase | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Grimshaw, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Tryptophan--tRNA ligase from Haemophilus influenzae.
to be published
|
|
6B8W
 
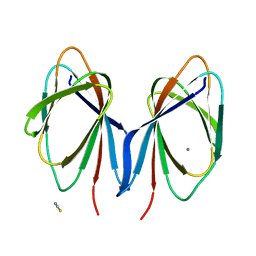 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6BNC
 
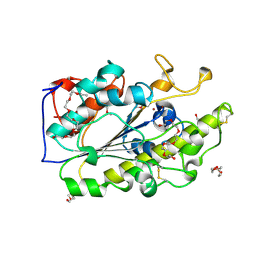 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant di-zinc and PEG complex | | Descriptor: | CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6BNF
 
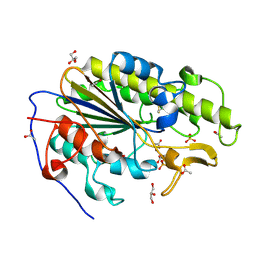 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
4ERR
 
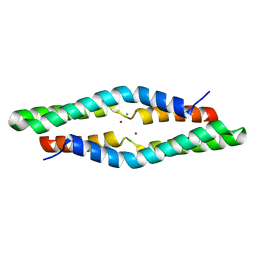 | | 1.55 Angstrom Crystal Structure of the Four Helical Bundle Membrane Localization Domain (4HBM) of the Vibrio vulnificus MARTX Effector Domain DUF5 | | Descriptor: | Autotransporter adhesin, BROMIDE ION, CHLORIDE ION | | Authors: | Minasov, G, Wawrzak, Z, Geissler, B, Shuvalova, L, Dubrovska, I, Winsor, J, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of the Four Helical Bundle Membrane Localization Domain (4HBM) of the Vibrio vulnificus MARTX Effector Domain DUF5.
TO BE PUBLISHED
|
|
1GSV
 
 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1HCJ
 
 | |
1GSW
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1ODV
 
 | | Photoactive yellow protein 1-25 deletion mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Vreede, J, Van Der horst, M.A, Hellingwerf, K.J, Crielaard, W, Van Aalten, D.M.F. | | Deposit date: | 2003-03-14 | | Release date: | 2003-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pas Domains.Common Structure and Common Flexibility
J.Biol.Chem., 278, 2003
|
|
2BUN
 
 | | Solution structure of the BLUF domain of AppA 5-125 | | Descriptor: | APPA, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Grinstead, J.S, Hsu, S.-T, Laan, W, Bonvin, A.M.J.J, Hellingwerf, K.J, Boelens, R, Kaptein, R. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the AppA BLUF domain: insight into the mechanism of light-induced signaling.
Chembiochem, 7, 2006
|
|
6WOP
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii | | Descriptor: | 4-aminobutyrate transaminase, CHLORIDE ION, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Acinetobacter baumannii
To Be Published
|
|
6WJ8
 
 | | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP | | Descriptor: | 4-aminobutyrate aminotransferase PuuE | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of gamma-aminobutyrate aminotransferase PuuE from Klebsiella pneumoniae in complex with PLP
To Be Published
|
|
