7BIM
 
 | | A de novo designed nonameric coiled coil, CC-Type2-(GgLaId)4 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Nonameric de novo coiled coil CC-Type2-(GgLaId)4 | | Authors: | Shelley, K.L, Rhys, G.G, Dawson, W.M, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Coiled coils 9-to-5: rational de novo design of alpha-helical barrels with tunable oligomeric states.
Chem Sci, 12, 2021
|
|
3JSQ
 
 | | Crystal structure of adipocyte fatty acid binding protein non-covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, Adipocyte fatty acid-binding protein, CHLORIDE ION, ... | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3JS1
 
 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
2GUZ
 
 | | Structure of the Tim14-Tim16 complex of the mitochondrial protein import motor | | Descriptor: | CITRATE ANION, Mitochondrial import inner membrane translocase subunit TIM14, Mitochondrial import inner membrane translocase subunit TIM16 | | Authors: | Mokranjac, D, Bourenkov, G, Hell, K, Neupert, W, Groll, M. | | Deposit date: | 2006-05-02 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor.
Embo J., 25, 2006
|
|
6UAP
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.745 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD6309 bound
To be Published
|
|
6U4X
 
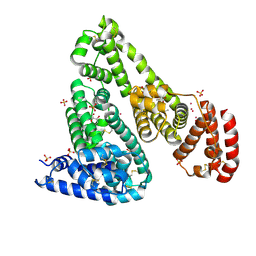 | | Crystal structure of Equine Serum Albumin complex with ibuprofen | | Descriptor: | IBUPROFEN, SULFATE ION, Serum albumin, ... | | Authors: | Czub, M.P, Handing, K.B, Venkataramany, B.S, Steen, E.H, Shabalin, I.G, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-26 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 63, 2020
|
|
6U69
 
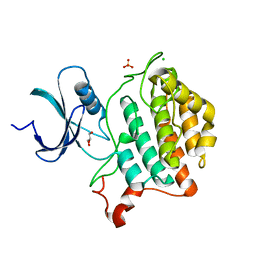 | | Crystal structure of Yck2 from Candida albicans, apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
7MQ5
 
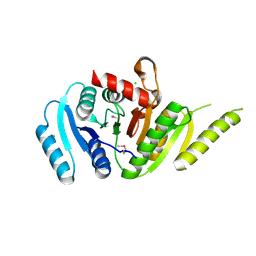 | | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14 | | Descriptor: | CHLORIDE ION, Universal stress protein | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Putative Universal Stress Protein from Pseudomonas aeruginosa UCBPP-PA14
To Be Published
|
|
4WBD
 
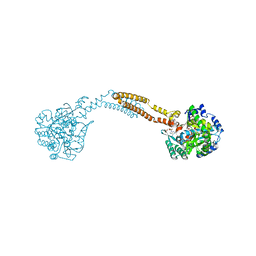 | | The crystal structure of BshC from Bacillus subtilis complexed with citrate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BSHC, CITRIC ACID, ... | | Authors: | Cook, P.D, VanDuinen, A.J, Winchell, K.R. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | X-ray Crystallographic Structure of BshC, a Unique Enzyme Involved in Bacillithiol Biosynthesis.
Biochemistry, 54, 2015
|
|
7N1M
 
 | | Crystal Structure of the Class D Beta-lactamase OXA-935 from Pseudomonas aeruginosa, Orthorhombic Crystal Form | | Descriptor: | Beta-lactamase OXA-935, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.B, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Antimicrob.Agents Chemother., 66, 2022
|
|
6OAD
 
 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
7MX5
 
 | | Crystal structure of TolB from Acinetobacter baumannii | | Descriptor: | Tol-Pal system protein TolB | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of TolB from Acinetobacter baumannii
To Be Published
|
|
7MWA
 
 | | Crystal structure of 2-octaprenyl-6-methoxyphenol hydroxylase UbiH from Acinetobacter baumannii, apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, 2-polyprenyl-6-methoxyphenol 4-hydroxylase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | UbiH from Acinetobacter baumannii
To Be Published
|
|
6PO4
 
 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
6UB9
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM | | Descriptor: | (2R,3S,4R)-3-(4'-chloro-2',6'-difluoro[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - AMINOACRYLATE- AND BRD6309-BOUND FORM
To be Published
|
|
6XKM
 
 | | Room Temperature Structure of SARS-CoV-2 NSP10/NSP16 Methyltransferase in a Complex with SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, CHLORIDE ION, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
6PNV
 
 | | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica | | Descriptor: | POTASSIUM ION, SODIUM ION, Tol-Pal system protein TolB | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica
To Be Published
|
|
6WJI
 
 | | 2.05 Angstrom Resolution Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2 | | Descriptor: | CHLORIDE ION, Nucleoprotein | | Authors: | Minasov, G, Shuvalova, L, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-13 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Serodominant SARS-CoV-2 Nucleocapsid Peptides Map to Unstructured Protein Regions.
Microbiol Spectr, 2023
|
|
8SFG
 
 | | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6 | | Descriptor: | Autotransporter adhesin, CHLORIDE ION, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Herrera, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-04-11 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Open Unbound Catalytically Inactive Makes Caterpillars Floppy-like (MCF) Effector from Vibrio vulnificus CMCP6.
To Be Published
|
|
