4AVZ
 
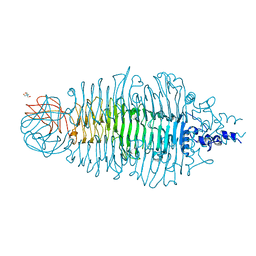 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 | | Descriptor: | TAIL SPIKE PROTEIN, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM | | Authors: | Gohlke, U, Broeker, N.K, Mueller, J.J, Uetrecht, C, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Single Amino Acid Exchange in Bacteriophage Hk620 Tailspike Protein Results in Thousand-Fold Increase of its Oligosaccharide Affinity.
Glycobiology, 23, 2013
|
|
7NDH
 
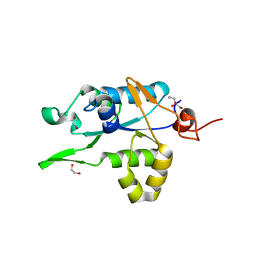 | | Crystal structure of ZC3H12C PIN domain | | Descriptor: | 1,2-ETHANEDIOL, Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDI
 
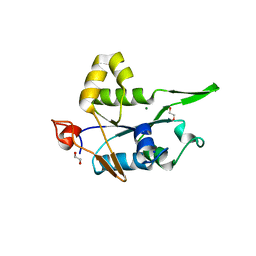 | | Crystal structure of ZC3H12C PIN domain with Mg2+ Ion | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable ribonuclease ZC3H12C, ... | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
7NDK
 
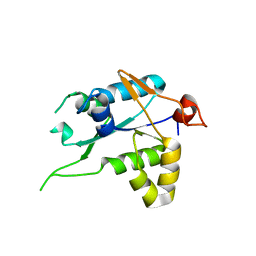 | | Crystal structure of ZC3H12C PIN catalytic mutant | | Descriptor: | Probable ribonuclease ZC3H12C, SODIUM ION | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PIN and CCCH Zn-finger domains coordinate RNA targeting in ZC3H12 family endoribonucleases.
Nucleic Acids Res., 49, 2021
|
|
2IW2
 
 | | Crystal structure of human Prolidase | | Descriptor: | SODIUM ION, XAA-PRO DIPEPTIDASE | | Authors: | Mueller, U, Niesen, F.H, Roske, Y, Goetz, F, Behlke, J, Buessow, K, Heinemann, U. | | Deposit date: | 2006-06-24 | | Release date: | 2006-07-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of Human Prolidase: The Molecular Basis of Pd Disease
To be Published
|
|
7OAS
 
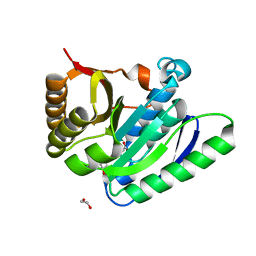 | | Structural basis for targeted p97 remodeling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine-dependent methyltransferases superfamily protein | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7OAT
 
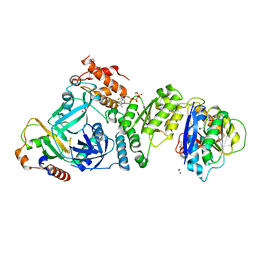 | | Structural basis for targeted p97 remodelling by ASPL as prerequisite for p97 trimethylation by METTL21D | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Petrovic, S, Heinemann, U, Roske, Y. | | Deposit date: | 2021-04-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.995 Å) | | Cite: | Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2XKW
 
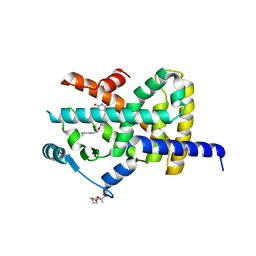 | | LIGAND BINDING DOMAIN OF HUMAN PPAR-GAMMA IN COMPLEX WITH THE AGONIST PIOGLITAZONE | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Mueller, J.J, Schupp, M, Unger, T, Kintscher, U, Heinemann, U. | | Deposit date: | 2010-07-14 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Binding Diversity of Pioglitazone by Peroxisome Proliferator-Activated Receptor-Gamma
To be Published
|
|
1GBG
 
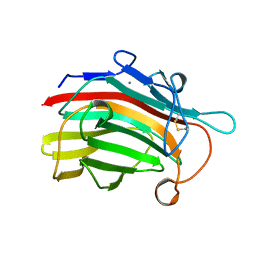 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
3ULJ
 
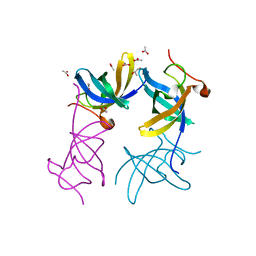 | | Crystal structure of apo Lin28B cold shock domain | | Descriptor: | ACETATE ION, GLYCEROL, Lin28b, ... | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-10 | | Release date: | 2012-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The Lin28 cold-shock domain remodels pre-let-7 microRNA.
Nucleic Acids Res., 40, 2012
|
|
1OJH
 
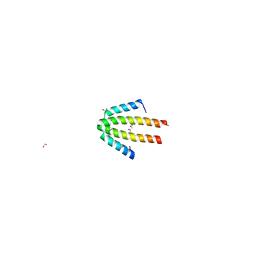 | | Crystal structure of NblA from PCC 7120 | | Descriptor: | 1,2-ETHANEDIOL, NBLA | | Authors: | Bienert, R, Baier, K, Lockau, W, Heinemann, U. | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Nbla from Anabaena Sp. Pcc 7120, a Small Protein Playing a Key Role in Phycobilisome Degradation.
J.Biol.Chem., 281, 2006
|
|
2JJZ
 
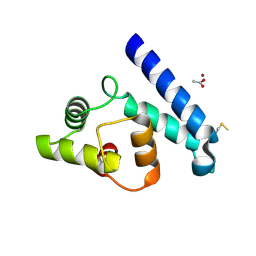 | | Crystal Structure of Human Iba2, orthorhombic crystal form | | Descriptor: | ACETATE ION, CHLORIDE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ... | | Authors: | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | Deposit date: | 2008-05-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
2XC1
 
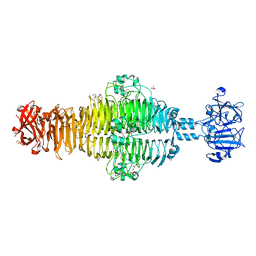 | | Full-length Tailspike Protein Mutant Y108W of Bacteriophage P22 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, ... | | Authors: | Mueller, J.J, Seul, A, Seckler, R, Heinemann, U. | | Deposit date: | 2010-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2YIL
 
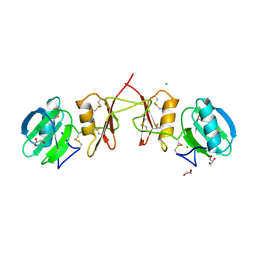 | | Crystal Structure of Parasite Sarcocystis muris Lectin SML-2 | | Descriptor: | CHLORIDE ION, GLYCEROL, MICRONEME ANTIGEN L2, ... | | Authors: | Mueller, J.J, Weiss, M.S, Heinemann, U. | | Deposit date: | 2011-05-16 | | Release date: | 2011-11-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-Modular Structure of Microneme Protein Sml-2 from Parasite Sarcocystis Muris at 1.95 A Resolution and its Complex with 1-Thio-Beta-D-Galactose.
Acta Crystallogr.,Sect.D, D67, 2011
|
|
2YIP
 
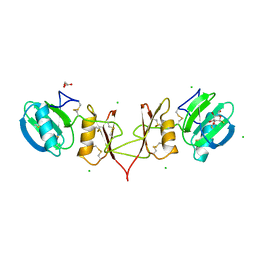 | |
5NXT
 
 | | Wobble base paired RNA double helix | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*UP*CP*UP*AP*CP*GP*AP*AP*GP*AP*AP*CP*A)-3') | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | A novel form of RNA double helix based on G·U and C·A+ wobble base pairing.
RNA, 24, 2018
|
|
1HZ9
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZA
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZB
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZC
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
3PR6
 
 | |
2BT6
 
 | | Ru(bpy)2(mbpy)-Modified Bovine Adrenodoxin | | Descriptor: | (4'-METHYL-2,2'BIPYRIDINE)BIS(2,2'-BIPYRIDINE), ADRENODOXIN 1, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Halavaty, A, Mueller, J.J, Contzen, J, Jung, C, Hannemann, F, Bernhardt, R, Galander, M, Lendzian, F, Heinemann, U. | | Deposit date: | 2005-05-26 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-Induced Reduction of Bovine Adrenodoxin Via the Covalently Bound Ruthenium(II) Bipyridyl Complex: Intramolecular Electron Transfer and Crystal Structure.
Biochemistry, 45, 2006
|
|
2CFH
 
 | | Structure of the Bet3-TPC6B core of TRAPP | | Descriptor: | PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 6B | | Authors: | Kummel, D, Muller, J.J, Roske, Y, Henke, N, Heinemann, U. | | Deposit date: | 2006-02-21 | | Release date: | 2006-07-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Bet3-Tpc6B Core of Trapp: Two Tpc6 Paralogs Form Trimeric Complexes with Bet3 and Mum2.
J.Mol.Biol., 361, 2006
|
|
1I5F
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD-SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-02-27 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1RGK
 
 | | RNASE T1 MUTANT GLU46GLN BINDS THE INHIBITORS 2'GMP AND 2'AMP AT THE 3' SUBSITE | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, CALCIUM ION, RIBONUCLEASE T1 | | Authors: | Granzin, J, Puras-Lutzke, R, Landt, O, Grunert, H.-P, Heinemann, U, Saenger, W, Hahn, U. | | Deposit date: | 1992-02-19 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | RNase T1 mutant Glu46Gln binds the inhibitors 2'GMP and 2'AMP at the 3' subsite.
J.Mol.Biol., 225, 1992
|
|
