2KDL
 
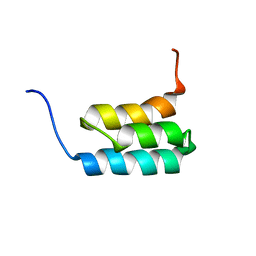 | | NMR structures of GA95 and GB95, two designed proteins with 95% sequence identity but different folds and functions | | Descriptor: | designed protein | | Authors: | He, Y, Alexander, P, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2009-01-12 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A minimal sequence code for switching protein structure and function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KQ8
 
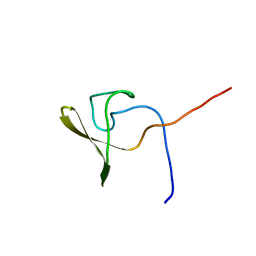 | | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A | | Descriptor: | Cell wall hydrolase | | Authors: | He, Y, Mills, J.L, Wu, Y, Eletsky, A, Wang, H, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a domain from BT9727_4915 from Bacillus thuringiensis, Northeast Structural Genomics Consortium Target BuR95A
To be Published
|
|
7YDM
 
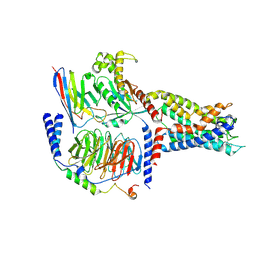 | | Cryo-EM structure of CD97/Gq complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
7YDP
 
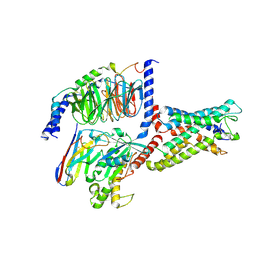 | | Cryo-EM structure of CD97/miniGs complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
7YDH
 
 | | Cryo EM structure of CD97/miniG13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, G protein subunit 13 (Gi2-mini-G13 chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of CD97 activation and G-protein coupling.
Cell Chem Biol, 30, 2023
|
|
2LHE
 
 | | Gb98-T25I,L20A | | Descriptor: | Gb98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2JWS
 
 | | Solution NMR structures of two designed proteins with 88% sequence identity but different fold and function | | Descriptor: | Ga88 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of two designed proteins with high sequence identity but different fold and function
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2LHG
 
 | | GB98-T25I solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHC
 
 | | Ga98 solution structure | | Descriptor: | Ga98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LHD
 
 | | GB98 solution structure | | Descriptor: | GB98 | | Authors: | He, Y, Chen, Y, Alexander, P, Bryan, P, Orban, J. | | Deposit date: | 2011-08-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mutational tipping points for switching protein folds and functions.
Structure, 20, 2012
|
|
2LU1
 
 | | pfsub2 solution NMR structure | | Descriptor: | Subtilase | | Authors: | He, Y, Chen, Y, Ruan, B, O'Brochta, D, Bryan, P, Orban, J. | | Deposit date: | 2012-06-06 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a sheddase inhibitor prodomain from the malarial parasite Plasmodium falciparum.
Proteins, 80, 2012
|
|
2MH8
 
 | | GA-79-MBP cs-rosetta structures | | Descriptor: | GA-79-MBP, maltose binding protein | | Authors: | He, Y, Chen, Y, Porter, L, Bryan, P, Orban, J. | | Deposit date: | 2013-11-19 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Subdomain interactions foster the design of two protein pairs with 80% sequence identity but different folds.
Biophys.J., 108, 2015
|
|
2MJY
 
 | | Solution structure of synthetic Mamba-1 peptide | | Descriptor: | Mambalgin-1 | | Authors: | He, Y, Wu, F, Tian, C. | | Deposit date: | 2014-01-21 | | Release date: | 2015-01-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | One-pot hydrazide-based native chemical ligation for efficient chemical synthesis and structure determination of toxin Mambalgin-1
Chem.Commun.(Camb.), 50, 2014
|
|
3R8W
 
 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
6LG2
 
 | | VanR bound to Vanillate | | Descriptor: | 4-HYDROXY-3-METHOXYBENZOATE, Predicted transcriptional regulators | | Authors: | He, Y, Bharath, S.R, Song, H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Developing a highly efficient hydroxytyrosol whole-cell catalyst by de-bottlenecking rate-limiting steps.
Nat Commun, 11, 2020
|
|
7WF7
 
 | | Cryo-EM of Sphingosine 1-phosphate receptor 1 / Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xu, Z. | | Deposit date: | 2021-12-26 | | Release date: | 2022-01-05 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of sphingosine-1-phosphate receptor 1 activation and biased agonism.
Nat.Chem.Biol., 18, 2022
|
|
7XV3
 
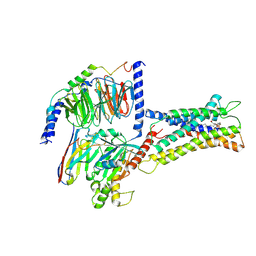 | | Cryo-EM structure of LPS-bound GPR174 in complex with Gs protein | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, Engineered G protein subunit S (mini-Gs), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Liang, J. | | Deposit date: | 2022-05-20 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation.
Nat Commun, 14, 2023
|
|
4P6X
 
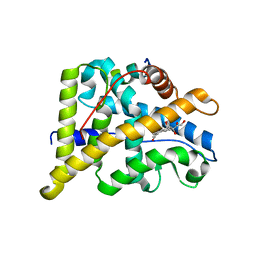 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
5XEY
 
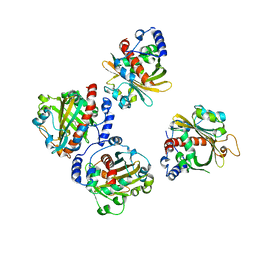 | | Discovery and structural analysis of a phloretin hydrolase from the opportunistic pathogen Mycobacterium abscessus | | Descriptor: | 1-[2,4,6-tris(oxidanyl)phenyl]ethanone, phloretin hydrolase | | Authors: | He, Y.X, Han, J.T, Jia, W.J, Zhang, Z. | | Deposit date: | 2017-04-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery and structural analysis of a phloretin hydrolase from the opportunistic human pathogen Mycobacterium abscessus.
FEBS J., 2019
|
|
5XTW
 
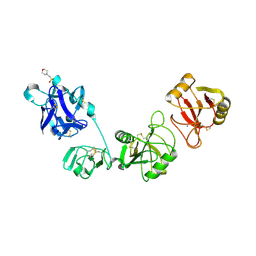 | | Crystal structure of the CysR-CTLD2 fragment of human MR at acidic pH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Macrophage mannose receptor 1 | | Authors: | He, Y, Hu, Z. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the pH-Dependent Conformational Change and Collagen Recognition of the Human Mannose Receptor
Structure, 26, 2018
|
|
5XTS
 
 | |
5XE5
 
 | | Discovery and structural analysis of a phloretin hydrolase from the opportunistic pathogen Mycobacterium abscessus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, phloretin hydrolase | | Authors: | He, Y.X, Han, J.T, Jia, W.J, Zhang, Z. | | Deposit date: | 2017-04-01 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and structural analysis of a phloretin hydrolase from the opportunistic human pathogen Mycobacterium abscessus.
FEBS J., 2019
|
|
2JU8
 
 | | Solution-State Structures of Oleate-Liganded LFABP, Major Form of 1:2 Protein-Ligand Complex | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | He, Y, Yang, X, Wang, H, Estephan, R, Francis, F, Kodukula, S, Storch, J, Stark, R.E. | | Deposit date: | 2007-08-15 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution-State Molecular Structure of Apo and Oleate-Liganded Liver Fatty Acid-Binding Protein
Biochemistry, 46, 2007
|
|
2JT4
 
 | |
4OIC
 
 | | Crystal structrual of a soluble protein | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Bet v I allergen-like, CHLORIDE ION, ... | | Authors: | He, Y, Hao, Q, Li, W, Yan, C, Yan, N, Yin, P. | | Deposit date: | 2014-01-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Identification and characterization of ABA receptors in Oryza sativa
Plos One, 9, 2014
|
|
