1CDQ
 
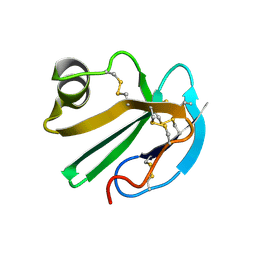 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
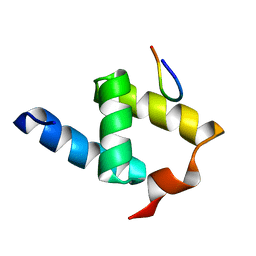
1OAI
 
 | |
2NPS
 
 | | Crystal Structure of the Early Endosomal SNARE Complex | | Descriptor: | Syntaxin 13, Syntaxin-6, Vesicle transport through interaction with t-SNAREs homolog 1A, ... | | Authors: | Zwilling, D, Cypionka, A, Pohl, W.H, Fasshauer, D, Walla, P.J, Wahl, M.C, Jahn, R. | | Deposit date: | 2006-10-30 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies
Embo J., 26, 2007
|
|
5TXS
 
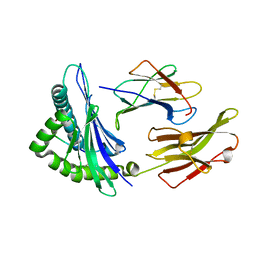 | | Crystal structure of an anaplastic lymphoma kinase-derived neuroblastoma tumor antigen bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-15 alpha chain, ... | | Authors: | Toor, J, Rao, A.A, Salama, S, Tripathi, S, Haussler, D, Sgourakis, N.G. | | Deposit date: | 2016-11-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | A Recurrent Mutation in Anaplastic Lymphoma Kinase with Distinct Neoepitope Conformations.
Front Immunol, 9, 2018
|
|
1XC5
 
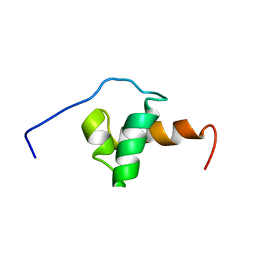 | | Solution Structure of the SMRT Deacetylase Activation Domain | | Descriptor: | Nuclear receptor corepressor 2 | | Authors: | Codina, A, Love, J.D, Li, Y, Lazar, M.A, Neuhaus, D, Schwabe, J.W.R. | | Deposit date: | 2004-09-01 | | Release date: | 2005-05-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2VRD
 
 | | THE STRUCTURE OF THE ZINC FINGER FROM THE HUMAN SPLICEOSOMAL PROTEIN U1C | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN C, ZINC ION | | Authors: | Muto, Y, Pomeranz-Krummel, D, Oubridge, C, Hernandez, H, Robinson, C, Neuhaus, D, Nagai, K. | | Deposit date: | 2008-03-31 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure and Biochemical Properties of the Human Spliceosomal Protein U1C
J.Mol.Biol., 341, 2004
|
|
1HCP
 
 | |
2MY1
 
 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2LBM
 
 | |
1XJR
 
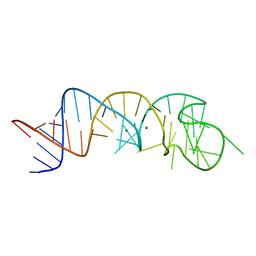 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | Descriptor: | MAGNESIUM ION, s2m RNA | | Authors: | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
2JM1
 
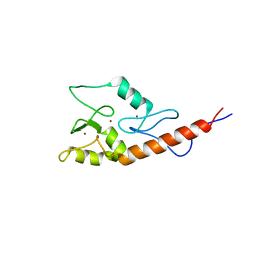 | |
1HF9
 
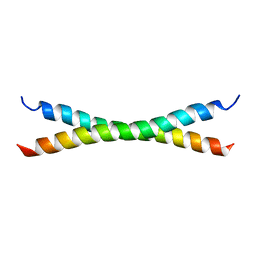 | | C-Terminal Coiled-Coil Domain from Bovine IF1 | | Descriptor: | ATPASE INHIBITOR (MITOCHONDRIAL) | | Authors: | Gordon-Smith, D.J, Carbajo, R.J, Yang, J.-C, Videler, H, Runswick, M.J, Walker, J.E, Neuhaus, D. | | Deposit date: | 2000-11-30 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase.
J. Mol. Biol., 308, 2001
|
|
1URQ
 
 | | Crystal structure of neuronal Q-SNAREs in complex with R-SNARE motif of Tomosyn | | Descriptor: | M-TOMOSYN ISOFORM, SYNAPTOSOMAL-ASSOCIATED PROTEIN 25, SYNTAXIN 1A | | Authors: | Pobbati, A, Razeto, A, Becker, S, Fasshauer, D. | | Deposit date: | 2003-10-31 | | Release date: | 2004-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibitory Role of Tomosyn in Exocytosis
J.Biol.Chem., 279, 2004
|
|
2JWQ
 
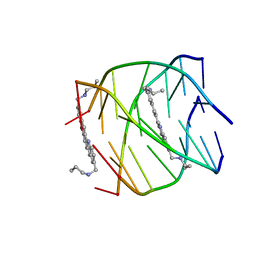 | | G-quadruplex recognition by quinacridines: a SAR, NMR and Biological study | | Descriptor: | DNA (5'-D(*DTP*DTP*DAP*DGP*DGP*DGP*DT)-3'), N,N'-(dibenzo[b,j][1,7]phenanthroline-2,10-diyldimethanediyl)dipropan-1-amine | | Authors: | Hounsou, C, Guittat, L, Monchaud, D, Jourdan, M, Saettel, N, Mergny, J.L, Teulade-Fichou, M. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | G-Quadruplex Recognition by Quinacridines: a SAR, NMR, and Biological Study
ChemMedChem, 2, 2007
|
|
2BN6
 
 | | P-Element Somatic Inhibitor Protein | | Descriptor: | PSI | | Authors: | Ignjatovic, T, Yang, J.C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BN5
 
 | | P-Element Somatic Inhibitor Protein Complex with U1-70k proline-rich peptide | | Descriptor: | PSI, U1 SMALL NUCLEAR RIBONUCLEOPROTEIN 70 KDA | | Authors: | Ignjatovic, T, Yang, J.-C, Butler, P.J.G, Neuhaus, D, Nagai, K. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction between P-Element Somatic Inhibitor and U1-70K Essential for the Alternative Splicing of P-Element Transposase.
J.Mol.Biol., 351, 2005
|
|
2BO5
 
 | | Bovine oligomycin sensitivity conferral protein N-terminal domain | | Descriptor: | ATP SYNTHASE OLIGOMYCIN SENSITIVITY CONFERRAL PROTEIN | | Authors: | Carbajo, R.J, Kellas, F.A, Runswick, M.J, Montgomery, M.G, Walker, J.E, Neuhaus, D. | | Deposit date: | 2005-04-07 | | Release date: | 2005-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit.
J. Mol. Biol., 351, 2005
|
|
3MSP
 
 | | MOTILE MAJOR SPERM PROTEIN (MSP) OF ASCARIS SUUM, NMR, 20 STRUCTURES | | Descriptor: | MAJOR SPERM PROTEIN | | Authors: | Haaf, A, Leclaire III, L, Roberts, G, Kent, H.M, Roberts, T.M, Stewart, M, Neuhaus, D. | | Deposit date: | 1998-09-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the motile major sperm protein (MSP) of Ascaris suum - evidence for two manganese binding sites and the possible role of divalent cations in filament formation.
J.Mol.Biol., 284, 1998
|
|
1SP7
 
 | | Structure of the Cys-rich C-terminal domain of Hydra minicollagen | | Descriptor: | mini-collagen | | Authors: | Meier, S, Haussinger, D, Pokidysheva, E, Bachinger, H.P, Grzesiek, S. | | Deposit date: | 2004-03-16 | | Release date: | 2004-05-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Determination of a high-precision NMR structure of the minicollagen cysteine rich domain from Hydra and characterization of its disulfide bond formation.
Febs Lett., 569, 2004
|
|
3ZNR
 
 | | HDAC7 bound with inhibitor TMP269 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[4-(4-phenyl-1,3-thiazol-2-yl)tetrahydro-2H-pyran-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, m, madauss, k, pohlhaus, d, trump, r, nolan, m. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
1UTA
 
 | | Solution structure of the C-terminal RNP domain from the divisome protein FtsN | | Descriptor: | CELL DIVISION PROTEIN FTSN | | Authors: | Yang, J.-C, van den Ent, F, Neuhaus, D, Brevier, J, Lowe, J. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Domain Architecture of the Divisome Protein Ftsn
Mol.Microbiol., 52, 2004
|
|
2K0A
 
 | | 1H, 15N and 13C chemical shift assignments for Rds3 protein | | Descriptor: | Pre-mRNA-splicing factor RDS3, ZINC ION | | Authors: | Loening, N, van Roon, A, Yang, J, Nagai, K, Neuhaus, D. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1UW0
 
 | |
2LHN
 
 | | RNA-binding zinc finger protein | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, ZINC ION | | Authors: | Brockmann, C, Neuhaus, D, Stewart, M. | | Deposit date: | 2011-08-12 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Polyadenosine-RNA Binding by Nab2 Zn Fingers and Its Function in mRNA Nuclear Export.
Structure, 20, 2012
|
|
1FHT
 
 | | RNA-BINDING DOMAIN OF THE U1A SPLICEOSOMAL PROTEIN U1A117, NMR, 43 STRUCTURES | | Descriptor: | U1 SMALL NUCLEAR RIBONUCLEOPROTEIN A | | Authors: | Allain, F.H.-T, Gubser, C.C, Howe, P.W.A, Nagai, K, Neuhaus, D, Varani, G. | | Deposit date: | 1996-02-21 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding.
J.Mol.Biol., 257, 1996
|
|
