6XYO
 
 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
2PJZ
 
 | | The crystal structure of putative Cobalt transport ATP-binding protein (cbiO-2), ST1066 | | Descriptor: | Hypothetical protein ST1066, SULFATE ION | | Authors: | Hirata, K, Hasegawa, K, Ebihara, A, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-17 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of putative Cobalt transport ATP-binding protein (cbiO-2), ST1066
To be Published
|
|
6LKN
 
 | | Crystal structure of ATP11C-CDC50A in PtdSer-bound E2P state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Hasegawa, K. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of a human plasma membrane phospholipid flippase.
J.Biol.Chem., 295, 2020
|
|
9ARQ
 
 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ARS
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9ART
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
1QPK
 
 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
6J8M
 
 | | Low-dose structure of bovine heart cytochrome c oxidase in the fully oxidized state determined using 30 keV X-ray | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Ueno, G, Shimada, A, Yamashita, E, Hasegawa, K, Kumasaka, T, Shinzawa-Itoh, K, Yoshikawa, S, Tsukihara, T, Yamamoto, M. | | Deposit date: | 2019-01-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Low-dose X-ray structure analysis of cytochrome c oxidase utilizing high-energy X-rays.
J.Synchrotron Radiat., 26, 2019
|
|
8K9N
 
 | | Subatomic resolution structure of Pseudoazurin from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Pseudoazurin, SULFATE ION | | Authors: | Fukuda, Y, Lintuluoto, M, Kurihara, K, Hasegawa, K, Inoue, T, Tamada, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Overlooked Hydrogen Bond in a Blue Copper Protein Uncovered by Neutron and Sub- angstrom ngstrom Resolution X-ray Crystallography.
Biochemistry, 63, 2024
|
|
8J5B
 
 | |
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
7UE1
 
 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | Authors: | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDA
 
 | | MALTOTETRAOSE-FORMING EXO-AMYLASE | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
5XAB
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2(TG) crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XAA
 
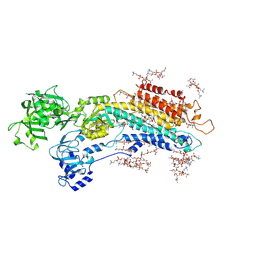 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of P21212 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA8
 
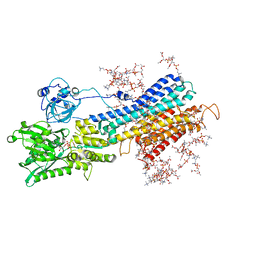 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-ALF4-ADP-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA9
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E2-ALF-(TG) crystals of C2 symmetry | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
5XA7
 
 | | Complete structure factors and an atomic model of the calcium pump (SERCA1A) and associated phospholipids in the E1-2CA2+ crystals | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Norimatsu, Y, Hasegawa, K, Shimizu, N, Toyoshima, C. | | Deposit date: | 2017-03-11 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Protein-phospholipid interplay revealed with crystals of a calcium pump.
Nature, 545, 2017
|
|
3VUO
 
 | | Crystal structure of nontoxic nonhemagglutinin subcomponent (NTNHA) from clostridium botulinum serotype D strain 4947 | | Descriptor: | NTNHA | | Authors: | Sagane, Y, Miyashita, S.-I, Miyata, K, Matsumoto, T, Inui, K, Hayashi, S, Suzuki, T, Hasegawa, K, Yajima, S, Yamano, A, Niwa, K, Watanabe, T. | | Deposit date: | 2012-07-03 | | Release date: | 2012-09-19 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Small-angle X-ray scattering reveals structural dynamics of the botulinum neurotoxin associating protein, nontoxic nonhemagglutinin
Biochem.Biophys.Res.Commun., 425, 2012
|
|
3AJN
 
 | | Structural basis of glycine amide on suppression of protein aggregation by high resolution X-ray analysis | | Descriptor: | AMINOMETHYLAMIDE, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Kumasaka, T. | | Deposit date: | 2010-06-09 | | Release date: | 2011-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Glycine amide shielding on the aromatic surfaces of lysozyme: Implication for suppression of protein aggregation
Febs Lett., 585, 2011
|
|
2AMG
 
 | | STRUCTURE OF HYDROLASE (GLYCOSIDASE) | | Descriptor: | 1,4-ALPHA-D-GLUCAN MALTOTETRAHYDROLASE, CALCIUM ION | | Authors: | Morishita, Y, Hasegawa, K, Matsuura, Y, Kubota, M, Sakai, S, Katsube, Y. | | Deposit date: | 1996-12-23 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a maltotetraose-forming exo-amylase from Pseudomonas stutzeri.
J.Mol.Biol., 267, 1997
|
|
3AGH
 
 | | X-ray analysis of lysozyme in the presence of 200 mM Arg | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Ito, L, Shiraki, K, Hasegawa, K, Baba, S, Kumasaka, T. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: implication of suppression of protein aggregation by arginine
Protein Eng.Des.Sel., 24, 2011
|
|
