2TBV
 
 | |
3N0A
 
 | | Crystal structure of auxilin (40-400) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein phosphatase auxilin | | Authors: | Harrison, S.C, Guan, R, Dai, H, Kirchhausen, T. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTEN-like Region of Auxilin, a Detector of Clathrin-Coated Vesicle Budding.
Structure, 18, 2010
|
|
1UZG
 
 | | CRYSTAL STRUCTURE OF THE DENGUE TYPE 3 VIRUS ENVELOPE PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAJOR ENVELOPE PROTEIN E, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modis, Y, Harrison, S.C. | | Deposit date: | 2004-03-11 | | Release date: | 2005-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein
J.Virol., 79, 2005
|
|
6NUW
 
 | | Yeast Ctf19 complex | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-02-03 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | The structure of the Ctf19c/CCAN from budding yeast.
Elife, 8, 2019
|
|
8G0P
 
 | |
8G0Q
 
 | |
3T79
 
 | | Ndc10: a platform for inner kinetochore assembly in budding yeast | | Descriptor: | DNA (5'-D(P*AP*AP*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*TP*TP*AP*TP*AP*AP*AP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*TP*TP*TP*AP*TP*AP*AP*AP*AP*TP*T)-3'), ... | | Authors: | Cho, U.S, Harrison, S.C. | | Deposit date: | 2011-07-29 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6113 Å) | | Cite: | Ndc10 is a platform for inner kinetochore assembly in budding yeast.
Nat.Struct.Mol.Biol., 19, 2011
|
|
4V7Q
 
 | | Atomic model of an infectious rotavirus particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Core scaffold protein, ... | | Authors: | Settembre, E.C, Chen, J.Z, Dormitzer, P.R, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2010-05-13 | | Release date: | 2014-07-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic model of an infectious rotavirus particle.
Embo J., 30, 2011
|
|
7UMK
 
 | | Structure of vesicular stomatitis virus (helical reconstruction, 4.1 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7UML
 
 | | Structure of vesicular stomatitis virus (local reconstruction, 3.5 A resolution) | | Descriptor: | Matrix protein, Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Jenni, S, Horwitz, J.A, Bloyet, L.-M, Whelan, S.P.J, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Visualizing molecular interactions that determine assembly of a bullet-shaped vesicular stomatitis virus particle.
Nat Commun, 13, 2022
|
|
7UI0
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with HSV010-13 Fab | | Descriptor: | Envelope glycoprotein B, HSV10-13 Fab Heavy chain, HSV10-13 Light chain | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
7UHZ
 
 | | Post-fusion ectodomain of HSV-1 gB in complex with BMPC-23 Fab | | Descriptor: | BMPC-23 Fab Heavy chain, BMPC-23 Fab Light chain, Envelope glycoprotein B | | Authors: | Windsor, I.W, Kong, S.L, Garforth, S.J, Almo, S.C, Harrison, S.C. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A non-neutralizing glycoprotein B monoclonal antibody protects against herpes simplex virus disease in mice.
J.Clin.Invest., 133, 2023
|
|
5KTB
 
 | |
7SWO
 
 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7UMS
 
 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 in complex with antibody 41 - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Fab 41 heavy chain, ... | | Authors: | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
7UMT
 
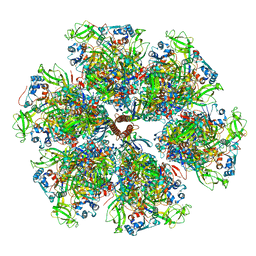 | | Structure of the VP5*/VP8* assembly from the human rotavirus strain CDC-9 - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Jenni, S, Zongli, L, Wang, Y, Bessey, T, Salgado, E.N, Schmidt, A.G, Greenberg, H.B, Jiang, B, Harrison, S.C. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotavirus VP4 Epitope of a Broadly Neutralizing Human Antibody Defined by Its Structure Bound with an Attenuated-Strain Virion.
J.Virol., 96, 2022
|
|
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8UDG
 
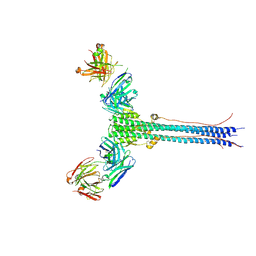 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8T0P
 
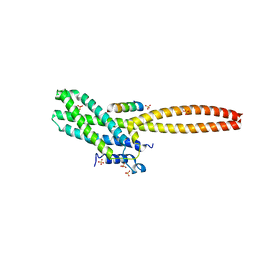 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
6Q0E
 
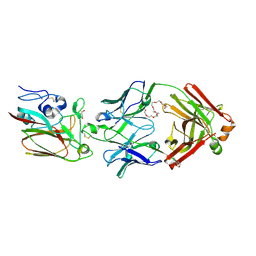 | |
6Q0O
 
 | |
7KDF
 
 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
