3N4S
 
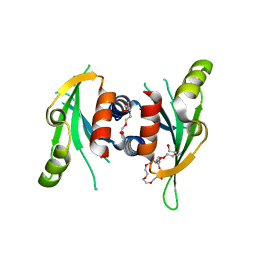 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
2IUH
 
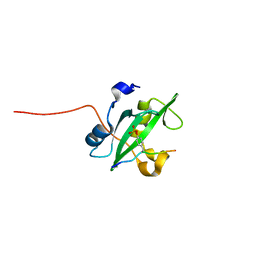 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with c-Kit phosphotyrosyl peptide | | Descriptor: | C-KIT PHOSPHOTYROSYL PEPTIDE, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUG
 
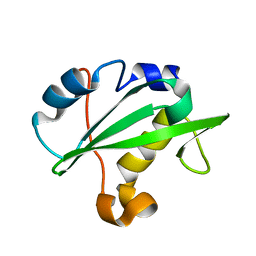 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY ALPHA SUBUNIT | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino-Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
2IUI
 
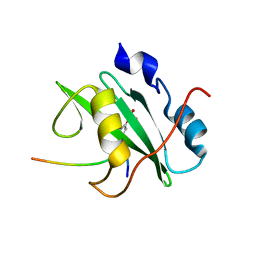 | | Crystal structure of the PI3-kinase p85 N-terminal SH2 domain in complex with PDGFR phosphotyrosyl peptide | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Platelet-derived growth factor receptor beta | | Authors: | Nolte, R.T, Eck, M.J, Schlessinger, J, Shoelson, S.E, Harrison, S.C. | | Deposit date: | 2006-06-03 | | Release date: | 2006-06-06 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Pi 3-Kinase P85 Amino- Terminal Sh2 Domain and its Phosphopeptide Complexes
Nat.Struct.Biol., 3, 1996
|
|
1KRI
 
 | |
1LCJ
 
 | |
1MOF
 
 | | COAT PROTEIN | | Descriptor: | CHLORIDE ION, MOLONEY MURINE LEUKEMIA VIRUS P15 | | Authors: | Fass, D, Harrison, S.C, Kim, P.S. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retrovirus envelope domain at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
5UIX
 
 | |
1BGW
 
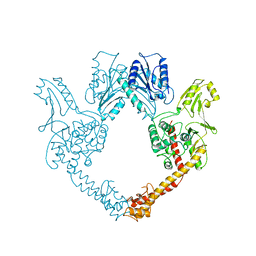 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
1SIE
 
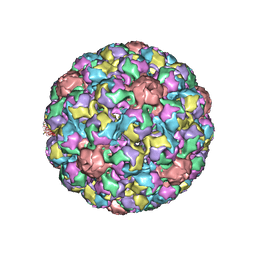 | | MURINE POLYOMAVIRUS COMPLEXED WITH A DISIALYLATED OLIGOSACCHARIDE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[N-acetyl-alpha-neuraminic acid-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, POLYOMAVIRUS COAT PROTEIN VP1 | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-06-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments.
Structure, 4, 1996
|
|
2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
2I3S
 
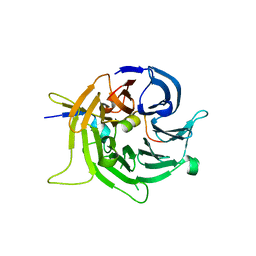 | | Bub3 complex with Bub1 GLEBS motif | | Descriptor: | Cell cycle arrest protein, Checkpoint serine/threonine-protein kinase | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2B6P
 
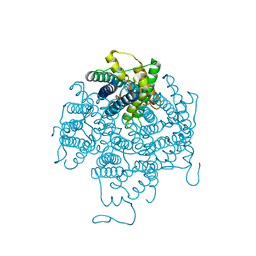 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
1OKE
 
 | |
2I3T
 
 | | Bub3 complex with Mad3 (BubR1) GLEBS motif | | Descriptor: | Cell cycle arrest protein, Spindle assembly checkpoint component | | Authors: | Larsen, N.A, Harrison, S.C. | | Deposit date: | 2006-08-20 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of Bub3 interactions in the mitotic spindle checkpoint.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1PYI
 
 | |
1SID
 
 | | MURINE POLYOMAVIRUS COMPLEXED WITH 3'SIALYL LACTOSE | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, POLYOMAVIRUS COAT PROTEIN VP1 | | Authors: | Stehle, T, Harrison, S.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments.
Structure, 4, 1996
|
|
1OAN
 
 | |
1SVA
 
 | | SIMIAN VIRUS 40 | | Descriptor: | SIMIAN VIRUS 40 | | Authors: | Stehle, T, Gamblin, S.J, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of simian virus 40 refined at 3.1 A resolution.
Structure, 4, 1996
|
|
1OK8
 
 | |
1TF6
 
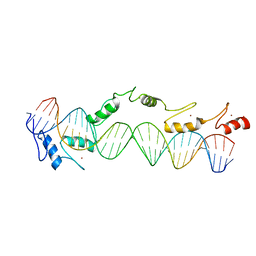 | | CO-CRYSTAL STRUCTURE OF XENOPUS TFIIIA ZINC FINGER DOMAIN BOUND TO THE 5S RIBOSOMAL RNA GENE INTERNAL CONTROL REGION | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*GP*CP*CP*TP*GP*GP*TP*TP*AP*GP*TP*AP*C P*CP*TP*GP*GP*AP* TP*GP*GP*GP*AP*GP*AP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*TP*CP*TP*CP*CP*CP*AP*TP*CP*CP*AP*GP*GP*T P*AP*CP*TP*AP*AP* CP*CP*AP*GP*GP*CP*CP*CP*G)-3'), PROTEIN (TRANSCRIPTION FACTOR IIIA), ... | | Authors: | Nolte, R.T, Conlin, R.M, Harrison, S.C, Brown, R.S. | | Deposit date: | 1998-03-02 | | Release date: | 1998-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Differing roles for zinc fingers in DNA recognition: structure of a six-finger transcription factor IIIA complex.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1R69
 
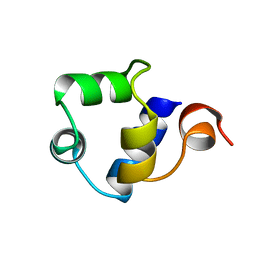 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
1LCK
 
 | |
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | Descriptor: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | Authors: | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | Deposit date: | 2005-05-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
