1EMW
 
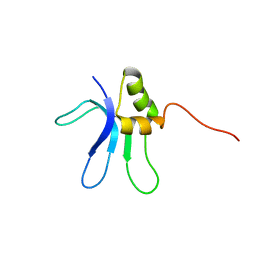 | | SOLUTION STRUCTURE OF THE RIBOSOMAL PROTEIN S16 FROM THERMUS THERMOPHILUS | | Descriptor: | S16 RIBOSOMAL PROTEIN | | Authors: | Allard, P, Rak, A.V, Wimberly, B.T, Clemons Jr, W.M, Kalinin, A, Helgstrand, M, Garber, M.B, Ramakrishnan, V, Hard, T. | | Deposit date: | 2000-03-20 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Another piece of the ribosome: solution structure of S16 and its location in the 30S subunit.
Structure Fold.Des., 8, 2000
|
|
1EAQ
 
 | | The RUNX1 Runt domain at 1.25A resolution: A structural switch and specifically bound chloride ions modulate DNA binding | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Backstrom, S, Wolf-Watz, M, Grundstrom, C, Hard, T, Grundstrom, T, Sauer, U.H. | | Deposit date: | 2001-07-14 | | Release date: | 2002-09-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The Runx1 Runt Domain at 1.25 A Resolution: A Structural Switch and Specifically Bound Chloride Ions Modulate DNA Binding
J.Mol.Biol., 322, 2002
|
|
1AB3
 
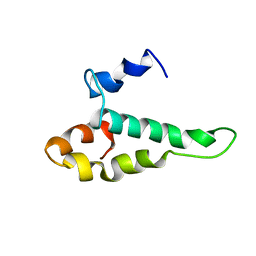 | | RIBOSOMAL PROTEIN S15 FROM THERMUS THERMOPHILUS, NMR, 26 STRUCTURES | | Descriptor: | RIBOSOMAL RNA BINDING PROTEIN S15 | | Authors: | Berglund, H, Rak, A, Serganov, A, Garber, M, Hard, T. | | Deposit date: | 1997-02-03 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ribosomal RNA binding protein S15 from Thermus thermophilus.
Nat.Struct.Biol., 4, 1997
|
|
2B89
 
 | |
2B88
 
 | |
2OTK
 
 | |
2B87
 
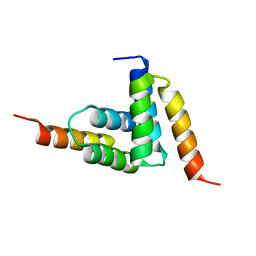 | |
4URK
 
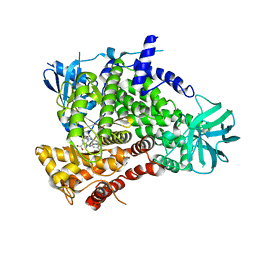 | | PI3Kg in complex with AZD6482 | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2QE4
 
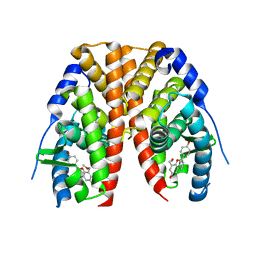 | | Estrogen receptor alpha ligand-binding domain in complex with a benzopyran agonist | | Descriptor: | (3AS,4R,9BR)-4-(4-HYDROXYPHENYL)-6-(METHOXYMETHYL)-1,2,3,3A,4,9B-HEXAHYDROCYCLOPENTA[C]CHROMEN-8-OL, Estrogen receptor | | Authors: | Norman, B.H, Richardson, T.I, Dodge, J.A, Pfeifer, L.A, Durst, G.L, Wang, Y, Durbin, J.D, Krishnan, V, Dinn, S.R, Liu, S.Q, Reilly, J.E, Ryter, K.T. | | Deposit date: | 2007-06-22 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Benzopyrans as selective estrogen receptor beta agonists (SERBAs). Part 4: Functionalization of the benzopyran A-ring.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3G04
 
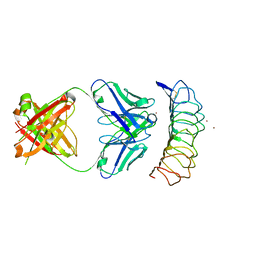 | | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN THYROID STIMULATING AUTOANTIBODY M22 HEAVY CHAIN, HUMAN THYROID STIMULATING AUTOANTIBODY M22 LIGHT CHAIN, ... | | Authors: | Sanders, J, Chirgadze, D.Y, Sanders, P, Baker, S, Sullivan, A, Bhardwaja, A, Bolton, J, Reeve, M, Nakatake, N, Evans, M, Richards, T, Powell, M, Miguel, R.N, Blundell, T.L, Furmaniak, J, Smith, B.R. | | Deposit date: | 2009-01-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody
Thyroid, 17, 2007
|
|
2PN9
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA modified aptamer | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*C)-3', RNA 16-mer with locked residues 9-10 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.-J. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
7NI3
 
 | | CRYSTAL STRUCTURE OF NATIVE HUMAN MYELOPEROXIDASE IN COMPLEX WITH CPD 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-sulfanylidene-3-[(2R)-tetrahydro-2-furanylmethyl]-1,2,3,7-tetrahydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Sjogren, T, Inghardt, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
J.Med.Chem., 65, 2022
|
|
7NI1
 
 | | CRYSTAL STRUCTURE OF NATIVE HUMAN MYELOPEROXIDASE IN COMPLEX WITH CPD 9 | | Descriptor: | (S)-1-(2-(amino(phenyl)methyl)benzyl)-2-thioxo-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sjogren, T, Inghardt, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
J.Med.Chem., 65, 2022
|
|
6Y6N
 
 | | Structure of mature activin A with small molecule 2 | | Descriptor: | (3~{R})-3,4-dimethyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
6BAR
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA (Q5SIX3_THET8) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Rod shape determining protein RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
6BAS
 
 | | Crystal structure of Thermus thermophilus Rod shape determining protein RodA D255A mutant (Q5SIX3_THET8) | | Descriptor: | CHLORIDE ION, Peptidoglycan glycosyltransferase RodA | | Authors: | Sjodt, M, Brock, K, Dobihal, G, Rohs, P.D.A, Green, A.G, Hopf, T.A, Meeske, A.J, Marks, D.S, Bernhardt, T.G, Rudner, D.Z, Kruse, A.C. | | Deposit date: | 2017-10-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis.
Nature, 556, 2018
|
|
7SQ2
 
 | | Reprocessed and refined structure of Phospholipase C-beta and Gq signaling complex | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, ACETATE ION, CALCIUM ION, ... | | Authors: | Endo-Streeter, S.T, Sondek, J, Harden, T.K. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Kinetic Scaffolding Mediated by a Phospholipase C-{beta} and Gq Signaling Complex
Science, 330, 2010
|
|
5ALB
 
 | | Ticagrelor antidote candidate MEDI2452 in complex with ticagrelor | | Descriptor: | MEDI2452 HEAVY CHAIN, MEDI2452 LIGHT CHAIN, Ticagrelor | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
5ALC
 
 | | Ticagrelor antidote candidate Fab 72 in complex with ticagrelor | | Descriptor: | ANTI-TICAGRELOR FAB 72, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
6Y6O
 
 | | Structure of mature activin A with small molecule 42 | | Descriptor: | (3~{R})-4-ethyl-3-methyl-3-propyl-1~{H}-1,4-benzodiazepine-2,5-dione, Inhibin beta A chain, SULFATE ION | | Authors: | McLoughlin, J.D, Brear, P.B, Reinhardt, T, Spring, D.R, Hyvonen, M. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
5F4N
 
 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | Authors: | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6PL6
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
4BH5
 
 | | LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Morlot, C, Peters, N.T, Yang, D.C, Uehara, T, Vernet, T, Bernhardt, T.G. | | Deposit date: | 2013-03-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-Function Analysis of the Lytm Domain of Envc, an Activator of Cell Wall Remodeling at the Escherichia Coli Division Site.
Mol.Microbiol., 89, 2013
|
|
2UW0
 
 | | Structure of PKA-PKB chimera complexed with 6-(4-(4-(4-Chloro-phenyl) -piperidin-4-yl)-phenyl)-9H-purine | | Descriptor: | 6-{4-[4-(4-CHLOROPHENYL)PIPERIDIN-4-YL]PHENYL}-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Hunter, L.J, Boyle, R.G, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2007-03-15 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rapid Evolution of 6-Phenylpurine Inhibitors of Protein Kinase B Through Structure-Based Design
J.Med.Chem., 50, 2007
|
|
