5X17
 
 | | Crystal structure of murine CK1d in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Casein kinase I isoform delta, SULFATE ION | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
3WBG
 
 | | Structure of the human heart fatty acid-binding protein in complex with 1-anilinonaphtalene-8-sulphonic acid | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, Fatty acid-binding protein, heart | | Authors: | Hirose, M, Sugiyama, S, Ishida, H, Niiyama, M, Matsuoka, D, Hara, T, Sato, F, Mizohata, E, Murakami, S, Inoue, T, Matsuoka, S, Murata, M. | | Deposit date: | 2013-05-16 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the human-heart fatty-acid-binding protein 3 in complex with the fluorescent probe 1-anilinonaphthalene-8-sulphonic acid
J.SYNCHROTRON RADIAT., 20, 2013
|
|
3WVM
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Matsumura, H, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type fatty-acid-binding protein.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1QH9
 
 | | ENZYME-PRODUCT COMPLEX OF L-2-HALOACID DEHALOGENASE | | Descriptor: | 2-HALOACID DEHALOGENASE, LACTIC ACID | | Authors: | Li, Y.-F, Hata, Y, Fujii, T, Kurihara, T, Esaki, N. | | Deposit date: | 1999-05-12 | | Release date: | 2000-05-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of L-2-Haloacid Dehalogenase Complexed with a Reaction Product Reveals the Mechanism of Intermediate Hydrolysis in Dehalogenase
To be Published
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
1QVC
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
4N2X
 
 | | Crystal Structure of DL-2-haloacid dehalogenase | | Descriptor: | DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
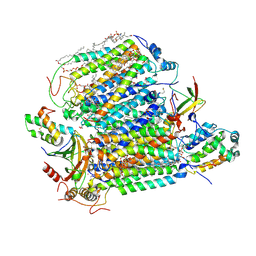 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7X73
 
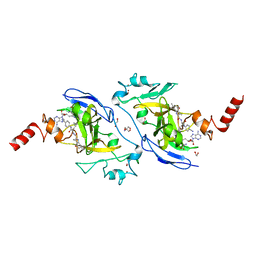 | | Structure of G9a in complex with RK-701 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nat Commun, 14, 2023
|
|
6M4W
 
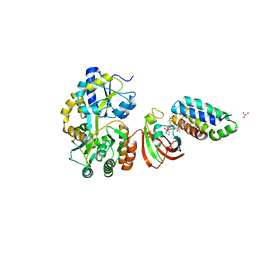 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
6TUQ
 
 | |
6TUP
 
 | |
5WRL
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y628 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
8YWO
 
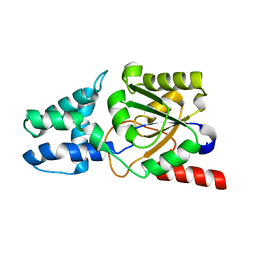 | | Crystal structure of L-azetidine-2-carboxylate hydrolase soaked in (S)-azetidine-2-carboxylic acid | | Descriptor: | (2S)-azetidine-2-carboxylic acid, (S)-2-haloacid dehalogenase | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-31 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
8YVW
 
 | | Crystal structure of D12N mutant of L-azetidine-2-carboxylate hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, FORMIC ACID, IMIDAZOLE, ... | | Authors: | Toyoda, M, Mizutani, K, Mikami, B, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2024-03-29 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Research for the crystal structure of L-azetidine-2-carboxylate hydrolase
To Be Published
|
|
2ELJ
 
 | | Solution structure of the SWIRM domain of baker's yeast Transcriptional adapter 2 | | Descriptor: | Transcriptional adapter 2 | | Authors: | Yoneyama, M, Tochio, N, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIRM domain of baker's yeast Transcriptional adapter 2
To be Published
|
|
2ELH
 
 | | Solution structure of the CENP-B N-terminal DNA-binding domain of fruit fly distal antenna CG11849-PA | | Descriptor: | CG11849-PA | | Authors: | Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CENP-B N-terminal DNA-binding domain of fruit fly distal antenna CG11849-PA
To be Published
|
|
2ELV
 
 | | Solution structure of the 6th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Tomizawa, T, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 6th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
2ELR
 
 | | Solution structure of the 15th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 15th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
2ELO
 
 | | Solution structure of the 12th C2H2 zinc finger of human Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 12th C2H2 zinc finger of human Zinc finger protein 406
To be Published
|
|
2ELW
 
 | | Solution structure of the 5th C2H2 zinc finger of mouse Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5th C2H2 zinc finger of mouse Zinc finger protein 406
To be Published
|
|
