6A83
 
 | |
6KRX
 
 | | Crystal Structure of AtPTP1 at 1.7 angstrom | | Descriptor: | CITRATE ANION, IODIDE ION, Protein-tyrosine-phosphatase PTP1 | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | AtPTP1 positively mediates brassinosteroid signaling from receptor kinases to GSK3-like kinase BIN2
To Be Published
|
|
6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
5YKI
 
 | | Crystal structure of the engineered nine-repeat PUF domain in complex with cognate 9nt-RNA | | Descriptor: | Pumilio homolog 1, RNA (5'-R(*UP*GP*UP*UP*GP*UP*AP*UP*A)-3') | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
5YKH
 
 | | Crystal structure of the engineered nine-repeat PUF domain | | Descriptor: | PHOSPHATE ION, Pumilio homolog 1 | | Authors: | Zhao, Y.Y, Wang, J, Li, H.T, Wang, Z.X, Wu, J.W. | | Deposit date: | 2017-10-14 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.457 Å) | | Cite: | Expanding RNA binding specificity and affinity of engineered PUF domains.
Nucleic Acids Res., 46, 2018
|
|
4O8S
 
 | |
5ZZU
 
 | |
1XPV
 
 | | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris | | Descriptor: | hypothetical protein XCC2852 | | Authors: | Shao, Y, Acton, T.B, Liu, G, Ma, L, Shen, Y, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-09 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Northeast Structural Genomics Target Protein XcR50 from X. Campestris
To be Published
|
|
4ILL
 
 | |
5KK2
 
 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
6DB9
 
 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | Descriptor: | Fab-Heavy-Chain, Fab-Light-Chain, MAGNESIUM ION, ... | | Authors: | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | Descriptor: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DHP
 
 | | Crystal structure of MazF from Deinococcus radiodurans | | Descriptor: | Endoribonuclease MazF, SULFATE ION | | Authors: | Zhao, Y, Dai, J. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | MazEF Toxin-Antitoxin System-Mediated DNA Damage Stress Response in Deinococcus radiodurans.
Front Genet, 12, 2021
|
|
6JY5
 
 | | Structure of CsoS4B from Halothiobacillus neapolitanus | | Descriptor: | Unidentified carboxysome polypeptide | | Authors: | Zhao, Y.Y, Jiang, Y.L, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of pentameric shell protein CsoS4B of Halothiobacillus neapolitanus alpha-carboxysome.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
4ILR
 
 | |
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
7S1Y
 
 | | Cryo-EM structure of human NKCC1 K289NA492E bound with bumetanide | | Descriptor: | 3-(butylamino)-4-phenoxy-5-sulfamoylbenzoic acid, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Zhao, Y.X, Cao, E.h. | | Deposit date: | 2021-09-02 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide
Nat Commun, 13, 2022
|
|
7S1X
 
 | |
7S1Z
 
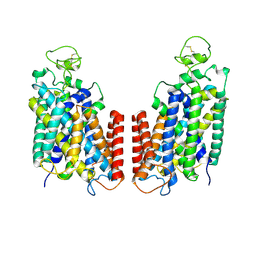 | | Cryo-EM structure of Human NKCC1 K289NA492EL671C | | Descriptor: | Solute carrier family 12 member 2 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2021-09-02 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for inhibition of the Cation-chloride cotransporter NKCC1 by the diuretic drug bumetanide
Nat Commun, 13, 2022
|
|
4H4K
 
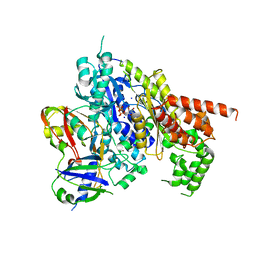 | | Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA-silencing complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Shao, Y, Cocozaki, A.I, Ramia, N.F, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2012-09-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Structure, 21, 2013
|
|
7TTI
 
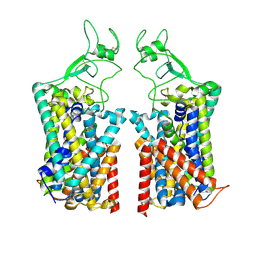 | | Human KCC1 bound with VU0463271 In an outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-cyclopropyl-N-(4-methyl-1,3-thiazol-2-yl)-2-[(6-phenylpyridazin-3-yl)sulfanyl]acetamide, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TTH
 
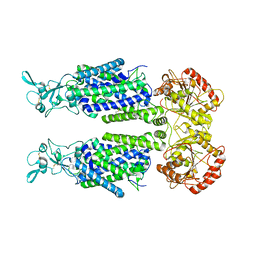 | | Human potassium-chloride cotransporter 1 in inward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, POTASSIUM ION, Solute carrier family 12 member 4 | | Authors: | Zhao, Y.X, Cao, E.H. | | Deposit date: | 2022-02-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structure of the human cation-chloride cotransport KCC1 in an outward-open state.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4ILM
 
 | | CRISPR RNA Processing endoribonuclease | | Descriptor: | CRISPR-associated endoribonuclease Cas6 2, RNA (5'-R(*GP*CP*UP*AP*AP*UP*CP*UP*AP*CP*UP*AP*UP*AP*GP*A)-3') | | Authors: | Shao, Y, Li, H. | | Deposit date: | 2012-12-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.068 Å) | | Cite: | Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6.
Structure, 21, 2013
|
|
5K4F
 
 | | CAL PDZ mutant C319A with a peptide | | Descriptor: | Golgi-associated PDZ and coiled-coil motif-containing protein, HPV18E6 peptide | | Authors: | Zhao, Y, Madden, D.R. | | Deposit date: | 2016-05-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CAL PDZ domain mutant with a peptide
To Be Published
|
|
