6ILM
 
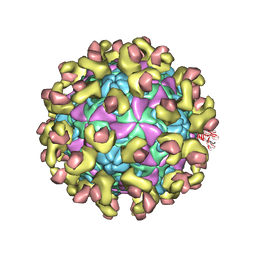 | | Cryo-EM structure of Echovirus 6 complexed with its uncoating receptor FcRn at PH 7.4 | | Descriptor: | Beta-2-microglobulin, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
6N0N
 
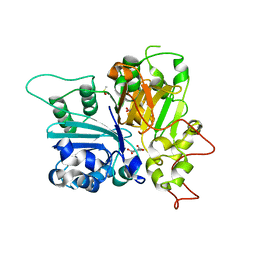 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ574 | | Descriptor: | 1,2-ETHANEDIOL, 4-methylbenzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6N0O
 
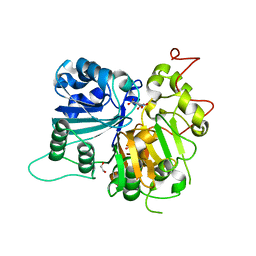 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ523 | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
6MYZ
 
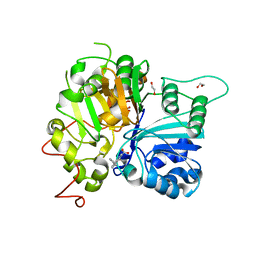 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ520 | | Descriptor: | 1,2-ETHANEDIOL, 4-oxo-8-phenyl-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Structure of Tdp1 catalytic domain in complex with compound XZ520
To Be Published
|
|
6N19
 
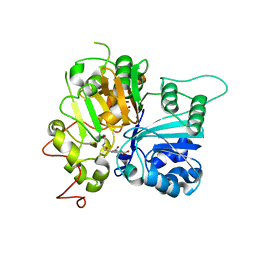 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ578 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-carboxybutanoyl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6ILO
 
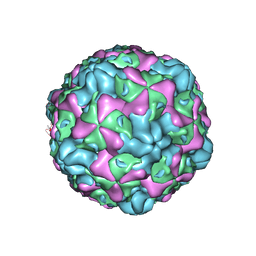 | | Cryo-EM structure of empty Echovirus 6 particle at PH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Gao, G.F, Liu, S, Zhao, X, Peng, R. | | Deposit date: | 2018-10-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human Neonatal Fc Receptor Is the Cellular Uncoating Receptor for Enterovirus B.
Cell, 177, 2019
|
|
2R25
 
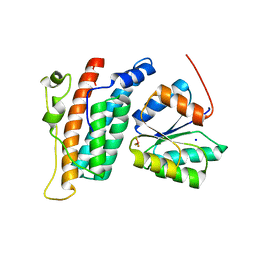 | | Complex of YPD1 and SLN1-R1 with bound Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Osmosensing histidine protein kinase SLN1, ... | | Authors: | Copeland, D.M, Zhao, X, Soares, A.S, West, A.H. | | Deposit date: | 2007-08-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a complex between the phosphorelay protein YPD1 and
the response regulator domain of SLN1 bound to a phosphoryl analog
J.Mol.Biol., 375, 2008
|
|
2D3D
 
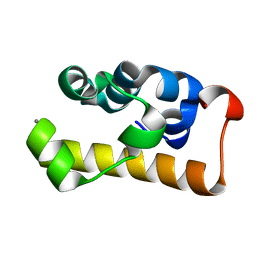 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
6DJH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ515 | | Descriptor: | 1,2-ETHANEDIOL, 8-bromo-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJG
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ503 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-sulfoquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
7DRV
 
 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
2IA8
 
 | | Kinetic and Crystallographic Studies of a Redesigned Manganese-Binding Site in Cytochrome c Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pfister, T, Mirarefi, A.Y, Gengenbach, A.J, Zhao, X, Conaster, C.D.N, Gao, Y.G, Robinson, H, Zukoski, C.F, Wang, A.H.J, Lu, Y. | | Deposit date: | 2006-09-07 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase
J.Biol.Inorg.Chem., 12, 2007
|
|
6DIH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
2ICV
 
 | | Kinetic and Crystallographic Studies of a Redesigned Manganese-Binding Site in Cytochrome c Peroxidase | | Descriptor: | COBALT (II) ION, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Pfister, T, Mirarefi, A.Y, Gengenbach, A.J, Zhao, X, Conaster, C.D.N, Gao, Y.G, Robinson, H, Zukoski, C.F, Wang, A.H.J, Lu, Y. | | Deposit date: | 2006-09-13 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase
J.Biol.Inorg.Chem., 12, 2007
|
|
5T7Q
 
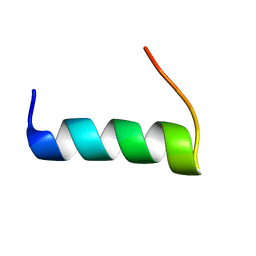 | | TIRAP phosphoinositide-binding motif | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Capelluto, D.G.S, Ellena, J.F, Armstrong, G, Zhao, X, Xiao, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane targeting of TIRAP is negatively regulated by phosphorylation in its phosphoinositide-binding motif.
Sci Rep, 7, 2017
|
|
6DJI
 
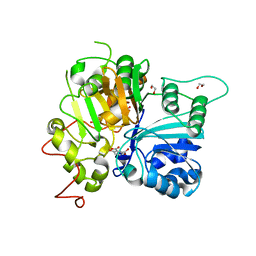 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ522 | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJJ
 
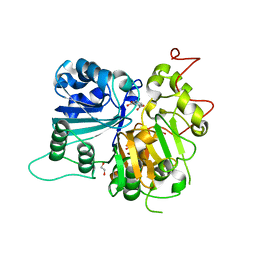 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
3RJX
 
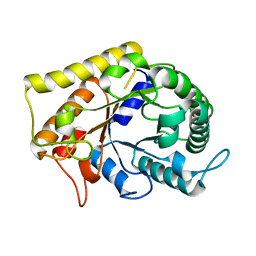 | | Crystal Structure of Hyperthermophilic Endo-Beta-1,4-glucanase | | Descriptor: | Endoglucanase FnCel5A | | Authors: | Zheng, B.S, Yang, W, Zhao, X.Y, Wang, Y.G, Lou, Z.Y, Rao, Z.H, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of hyperthermophilic endo-beta-1,4-glucanase: implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
3GD7
 
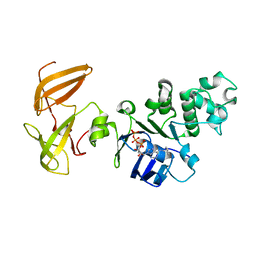 | | Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP) | | Descriptor: | Fusion complex of Cystic fibrosis transmembrane conductance regulator, residues 1193-1427 and Maltose/maltodextrin import ATP-binding protein malK, residues 219-371, ... | | Authors: | Atwell, S, Antonysamy, S, Conners, K, Emtage, S, Gheyi, T, Lewis, H.A, Lu, F, Sauder, J.M, Wasserman, S.R, Zhao, X. | | Deposit date: | 2009-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human NBD2 complexed with N6-Phenylethyl-ATP (P-ATP)
To be Published
|
|
1XMJ
 
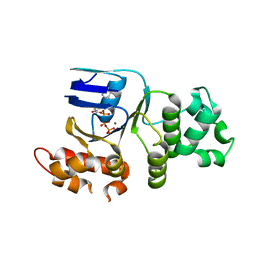 | | Crystal structure of human deltaF508 human NBD1 domain with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Zhao, X, Wang, C, Sauder, J.M, Rooney, I, Noland, B.W, Lorimer, D, Kearins, M.C, Conners, K, Condon, B, Maloney, P.C, Guggino, W.B, Hunt, J.F, Emtage, S, Structural GenomiX | | Deposit date: | 2004-10-02 | | Release date: | 2004-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Impact of the delta-F508 Mutation in First Nucleotide-binding Domain of Human Cystic Fibrosis Transmembrane Conductance Regulator on Domain Folding and Structure
J.Biol.Chem., 280, 2005
|
|
4C51
 
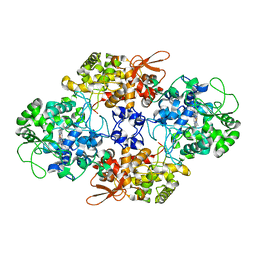 | | Crystal Structure of the Catalase-Peroxidase (KatG) R418L mutant from Mycobacterium Tuberculosis | | Descriptor: | CATALASE-PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, alpha-D-glucopyranose | | Authors: | Hersleth, H.-P, Zhao, X, Magliozzo, R.S, Andersson, K.K. | | Deposit date: | 2013-09-10 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Access Channel Residues Ser315 and Asp137 in Mycobacterium Tuberculosis Catalase-Peroxidase (Katg) Control Peroxidatic Activation of the Pro-Drug Isoniazid.
Chem.Commun.(Camb.), 49, 2013
|
|
4C50
 
 | | Crystal Structure of the Catalase-Peroxidase (KatG) D137S mutant from Mycobacterium Tuberculosis | | Descriptor: | ACETATE ION, CATALASE-PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hersleth, H.-P, Zhao, X, Magliozzo, R.S, Andersson, K.K. | | Deposit date: | 2013-09-10 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Access Channel Residues Ser315 and Asp137 in Mycobacterium Tuberculosis Catalase-Peroxidase (Katg) Control Peroxidatic Activation of the Pro-Drug Isoniazid.
Chem.Commun.(Camb.), 49, 2013
|
|
2BBO
 
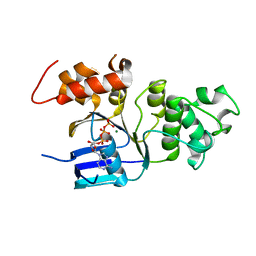 | | Human NBD1 with Phe508 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
2BBS
 
 | | Human deltaF508 NBD1 with three solubilizing mutations | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
2BBT
 
 | | Human deltaF508 NBD1 with two solublizing mutations. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Lewis, H.A, Kearins, M.C, Conners, K, Zhao, X, Lu, F, Sauder, J.M, Emtage, S. | | Deposit date: | 2005-10-17 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry.
J.Mol.Biol., 396, 2010
|
|
