7V5K
 
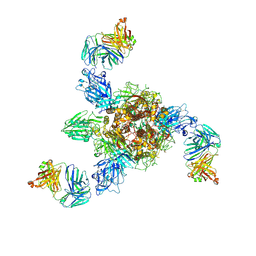 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1) | | 分子名称: | 0722 H, 0722 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-17 | | 公開日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 0722 (state 1)
to be published
|
|
7V5J
 
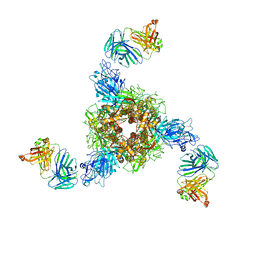 | | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2) | | 分子名称: | 0722 H, 0722 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-17 | | 公開日 | 2022-09-21 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 0722(state 2)
to be published
|
|
7V6N
 
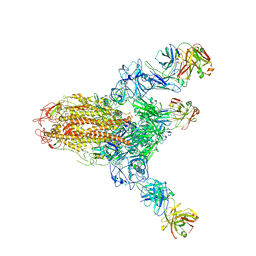 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | 分子名称: | 111 H, 111 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-20 | | 公開日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
7V6O
 
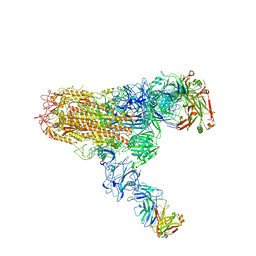 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2) | | 分子名称: | 111 H, 111 L, Spike glycoprotein | | 著者 | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | 登録日 | 2021-08-20 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.56 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2)
to be published
|
|
7V3L
 
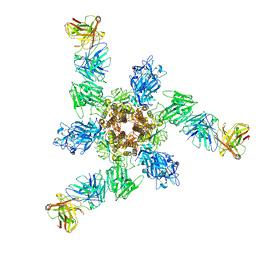 | | MERS S ectodomain trimer in complex with neutralizing antibody 6516 | | 分子名称: | Spike glycoprotein, antibody H, antibody L | | 著者 | Wang, X, Zhao, J, Wang, Z, Wang, Y, Zeng, J. | | 登録日 | 2021-08-10 | | 公開日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | MERS S ectodomain trimer in complex with neutralizing antibody 6516
to be published
|
|
7RUH
 
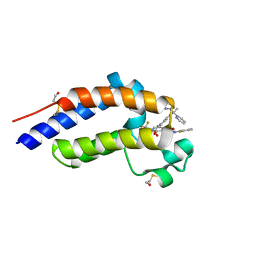 | | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844 | | 分子名称: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2021-08-17 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Bromodomain-containing protein 4 (BRD4) bromodomain 2 (BD2) complexed with XR844
To Be Published
|
|
7RUI
 
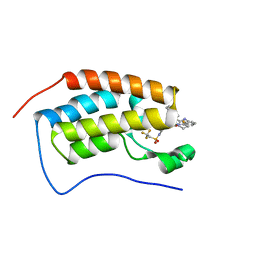 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844 | | 分子名称: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-(5-{[(2-fluorophenyl)carbamoyl]amino}-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-1H-indol-4-yl}-2,2,2-trifluoroethane-1-sulfonamide | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2021-08-17 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with XR844
To Be Published
|
|
7L6W
 
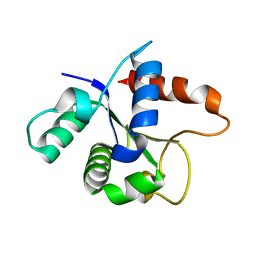 | | SFX structure of the MyD88 TIR domain higher-order assembly | | 分子名称: | Myeloid differentiation primary response protein MyD88 | | 著者 | Clabbers, M.T.B, Holmes, S, Muusse, T.W, Vajjhala, P, Thygesen, S.J, Malde, A.K, Hunter, D.J.B, Croll, T.I, Flueckiger, L, Nanson, J.D, Rahaman, M.H, Aquila, A, Hunter, M.S, Liang, M, Yoon, C.H, Zhao, J, Zatsepin, N.A, Abbey, B, Sierecki, E, Gambin, Y, Stacey, K.J, Darmanin, C, Kobe, B, Xu, H, Ve, T. | | 登録日 | 2020-12-24 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | MyD88 TIR domain higher-order assembly interactions revealed by microcrystal electron diffraction and serial femtosecond crystallography.
Nat Commun, 12, 2021
|
|
7JKX
 
 | | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6 | | 分子名称: | Bromodomain-containing protein 4, N-{1-[1,1-di(pyridin-2-yl)ethyl]-6-[1-methyl-5-(methylamino)-6-oxo-1,6-dihydropyridin-3-yl]-1H-indol-4-yl}ethanesulfonamide | | 著者 | Ratia, K.M, Xiong, R, Li, Y, Shen, Z, Zhao, J, Huang, F, Dubrovyskyii, O, Thatcher, G.R. | | 登録日 | 2020-07-29 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Bromodomain-containing protein 4 (BRD4) bromodomain 1 (BD1) complexed with YF3-6
To Be Published
|
|
5F0U
 
 | |
5GAS
 
 | | Thermus thermophilus V/A-ATPase, conformation 2 | | 分子名称: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | 著者 | Schep, D.G, Zhao, J, Rubinstein, J.L. | | 登録日 | 2016-02-05 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (9.5 Å) | | 主引用文献 | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4O98
 
 | | Crystal structure of Pseudomonas oleovorans PoOPH mutant H250I/I263W | | 分子名称: | ZINC ION, organophosphorus hydrolase | | 著者 | Luo, X.J, Kong, X.D, Zhao, J, Chen, Q, Zhou, J.H, Xu, J.H. | | 登録日 | 2014-01-02 | | 公開日 | 2014-12-03 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Switching a newly discovered lactonase into an efficient and thermostable phosphotriesterase by simple double mutations His250Ile/Ile263Trp
Biotechnol.Bioeng., 111, 2014
|
|
5GAR
 
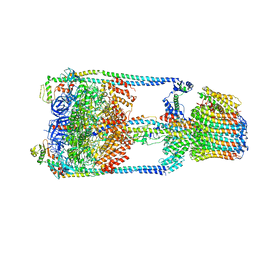 | | Thermus thermophilus V/A-ATPase, conformation 1 | | 分子名称: | Archaeal/vacuolar-type H+-ATPase subunit I, V-type ATP synthase alpha chain, V-type ATP synthase beta chain, ... | | 著者 | Schep, D.G, Zhao, J, Rubinstein, J.L. | | 登録日 | 2016-02-05 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3URM
 
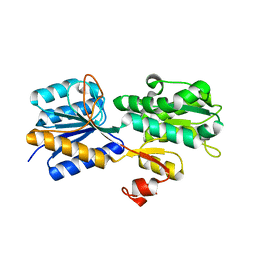 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-22 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5F0W
 
 | |
3UUG
 
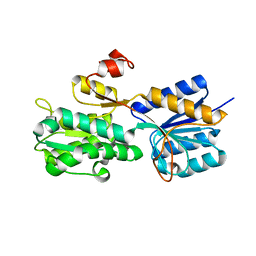 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5HXD
 
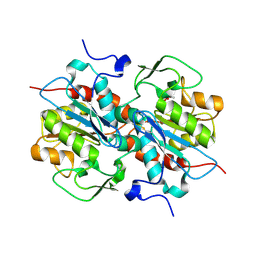 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | 分子名称: | CACODYLATE ION, Protein MpaA, ZINC ION | | 著者 | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | 登録日 | 2016-01-30 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
5I1M
 
 | |
1PSF
 
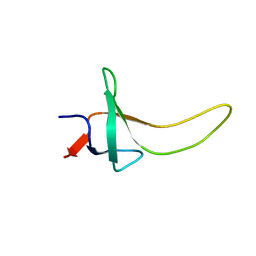 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF PSAE FROM THE CYANOBACTERIUM SYNECHOCOCCUS SP. STRAIN PCC 7002: A PHOTOSYSTEM I PROTEIN THAT SHOWS STRUCTURAL HOMOLOGY WITH SH3 DOMAINS | | 分子名称: | PHOTOSYSTEM I ACCESSORY PROTEIN E | | 著者 | Falzone, C.J, Kao, Y.-H, Zhao, J, Bryant, D.A, Lecomte, J.T.J. | | 登録日 | 1994-04-13 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of PsaE from the cyanobacterium Synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains.
Biochemistry, 33, 1994
|
|
1PSE
 
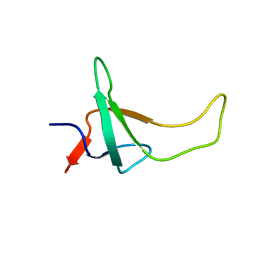 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF PSAE FROM THE CYANOBACTERIUM SYNECHOCOCCUS SP. STRAIN PCC 7002: A PHOTOSYSTEM I PROTEIN THAT SHOWS STRUCTURAL HOMOLOGY WITH SH3 DOMAINS | | 分子名称: | PHOTOSYSTEM I ACCESSORY PROTEIN E | | 著者 | Falzone, C.J, Kao, Y.-H, Zhao, J, Bryant, D.A, Lecomte, J.T.J. | | 登録日 | 1994-04-13 | | 公開日 | 1995-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of PsaE from the cyanobacterium Synechococcus sp. strain PCC 7002, a photosystem I protein that shows structural homology with SH3 domains.
Biochemistry, 33, 1994
|
|
6K61
 
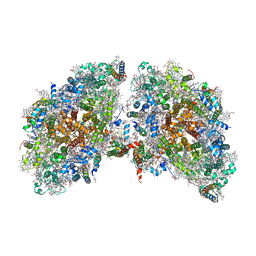 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | 登録日 | 2019-05-31 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.37 Å) | | 主引用文献 | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
4DWU
 
 | |
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | 分子名称: | COPPER (II) ION, PLASTOCYANIN | | 著者 | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | 登録日 | 2001-09-07 | | 公開日 | 2001-09-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
2N4H
 
 | |
1JXF
 
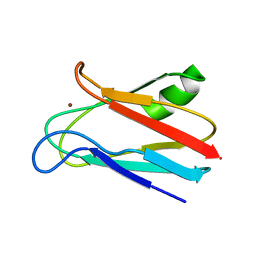 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | 分子名称: | COPPER (II) ION, PLASTOCYANIN | | 著者 | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | 登録日 | 2001-09-07 | | 公開日 | 2001-09-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
