3LID
 
 | |
3SSZ
 
 | | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium | | Descriptor: | Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | The crystal structure of Mandelate racemase/muconate lactonizing enzyme from Rhodobacteraceae bacterium
To be Published
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
3LNV
 
 | | The crystal structure of fatty acyl-adenylate ligase from L. pneumophila in complex with acyl adenylate and pyrophosphate | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, PYROPHOSPHATE 2-, Saframycin Mx1 synthetase B | | Authors: | Zhang, Z, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-03 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Studies of Fatty Acyl Adenylate Ligases from E. coli and L. pneumophila.
J.Mol.Biol., 406, 2011
|
|
3GBT
 
 | |
3JBQ
 
 | | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6 | | Descriptor: | GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, IgG1-kappa 2E8 heavy chain, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
6LVZ
 
 | | Crystal structure of TLR7/Cpd-3 (SM-394830) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(pyridin-3-ylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
3LIA
 
 | |
6LW0
 
 | | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purin-6-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
3LL3
 
 | |
6LVY
 
 | | Crystal structure of TLR7/Cpd-2 (SM-360320) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LR7
 
 | |
3LWI
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3LWH
 
 | | Crystal structure of Cren7-dsDNA complex | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Gong, Y, Guo, L, Jiang, T, Huang, L. | | Deposit date: | 2010-02-23 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA
Mol.Microbiol., 76, 2010
|
|
3TBL
 
 | | Structure of Mono-ubiquitinated PCNA: Implications for DNA Polymerase Switching and Okazaki Fragment Maturation | | Descriptor: | Proliferating cell nuclear antigen, Ubiquitin | | Authors: | Zhang, Z, Lee, M, Lee, E, Zhang, S. | | Deposit date: | 2011-08-07 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of monoubiquitinated PCNA: Implications for DNA polymerase switching and Okazaki fragment maturation.
Cell Cycle, 11, 2012
|
|
7YTP
 
 | | TLR7 in complex with an inhibitor | | Descriptor: | (2R,6R)-4-(8-cyanoquinolin-5-yl)-N-[(3S,4R)-4-fluoranylpyrrolidin-3-yl]-6-methyl-morpholine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2022-08-15 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | TLR7 in complex with an inhibitor
To Be Published
|
|
3M2P
 
 | |
3PZL
 
 | |
3MPO
 
 | |
3I8B
 
 | |
3NIV
 
 | |
3NND
 
 | |
2KKA
 
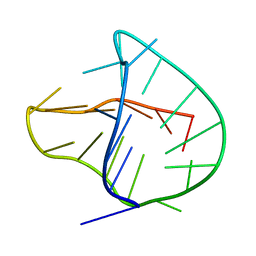 | | Human telomere DNA two-tetrad quadruplex structure in K+ solution | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*IP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3' | | Authors: | Zhang, Z, Dai, J, Yang, D. | | Deposit date: | 2009-06-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures.
Nucleic Acids Res., 38, 2010
|
|
3NPK
 
 | |
4LK2
 
 | | Crystal structure of RNA splicing effector Prp5 | | Descriptor: | NICKEL (II) ION, Pre-mRNA-processing ATP-dependent RNA helicase PRP5 | | Authors: | Zhang, Z.-M, Li, J, Yang, F, Xu, Y, Zhou, J. | | Deposit date: | 2013-07-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly.
Cell Rep, 5, 2013
|
|
