2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
7CG7
 
 | | Cryo-EM structure of the flagellar MS ring with C34 symmetry from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGB
 
 | | Cryo-EM structure of the flagellar hook from Salmonella | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBM
 
 | | Cryo-EM structure of the flagellar distal rod with partial hook from Salmonella | | Descriptor: | Flagellar basal-body rod protein FlgF, Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
3OS5
 
 | | SET7/9-Dnmt1 K142me1 complex | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Dnmt1, ... | | Authors: | Esteve, P.-O, Chang, Y, Samaranayake, M, Upadhyay, A.K, Horton, J.R, Feehery, G.R, Cheng, X, Pradhan, S. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7EV9
 
 | | cryoEM structure of particulate methane monooxygenase associated with Cu(I) | | Descriptor: | Ammonia monooxygenase/methane monooxygenase, subunit C family protein, COPPER (I) ION, ... | | Authors: | Chang, W.H, Lin, H.H, Tsai, I.K, Huang, S.H, Chung, S.C, Tu, I.P, Yu, S.F, Chan, S.I. | | Deposit date: | 2021-05-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Copper Centers in the Cryo-EM Structure of Particulate Methane Monooxygenase Reveal the Catalytic Machinery of Methane Oxidation.
J.Am.Chem.Soc., 143, 2021
|
|
1XBB
 
 | | Crystal structure of the syk tyrosine kinase domain with Gleevec | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Tyrosine-protein kinase SYK | | Authors: | Nienaber, V.L, Atwell, S, Adams, J.M, Badger, J, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Novel Mode of Gleevec Binding Is Revealed by the Structure of Spleen Tyrosine Kinase
J.Biol.Chem., 279, 2004
|
|
1XBC
 
 | | Crystal structure of the syk tyrosine kinase domain with Staurosporin | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase SYK | | Authors: | Badger, J, Atwell, S, Adams, J.M, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase
J.Biol.Chem., 279, 2004
|
|
7C31
 
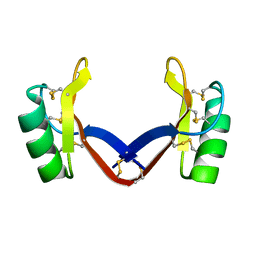 | | Crystal structure of the grapevine defensin VvK1 | | Descriptor: | Knot1 domain-containing protein | | Authors: | Chen, M.W, Chang, S.C, Chandy, K.G, Luo, D. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Modulation of Lymphocyte Potassium Channel KV1.3 by Membrane-Penetrating, Joint-Targeting Immunomodulatory Plant Defensin.
Acs Pharmacol Transl Sci, 3, 2020
|
|
1XBA
 
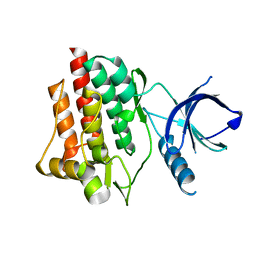 | | Crystal structure of apo syk tyrosine kinase domain | | Descriptor: | Tyrosine-protein kinase SYK | | Authors: | Atwell, S, Adams, J.M, Badger, J, Buchanan, M.D, Feil, I.K, Froning, K.J, Gao, X, Hendle, J, Keegan, K, Leon, B.C, Muller-Deickmann, H.J, Nienaber, V.L, Noland, B.W, Post, K, Rajashankar, K.R, Ramos, A, Russell, M, Burley, S.K, Buchanan, S.G. | | Deposit date: | 2004-08-30 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of Gleevec binding is revealed by the structure of spleen tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
2B1N
 
 | |
6PNJ
 
 | | Structure of Photosystem I Acclimated to Far-red Light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Shen, G, Kurashov, V, Ho, M, Zhang, S, Williams, D, Golbeck, J.H, Fromme, P, Bryant, D.A. | | Deposit date: | 2019-07-02 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structure of Photosystem I acclimated to far-red light illuminates an ecologically important acclimation process in photosynthesis
Sci Adv, 6, 2020
|
|
1XMA
 
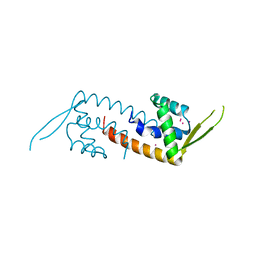 | | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833 | | Descriptor: | MERCURY (II) ION, Predicted transcriptional regulator, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Chen, L, Lee, D, Habel, J, Nguyen, J, Chang, S.-H, Kataeva, I, Xu, H, Chang, J, Zhao, M, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Praissman, J, Zhang, H, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-01 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of a transcriptional regulator from Clostridium thermocellum Cth-833
To be published
|
|
1XRG
 
 | | Conserved hypothetical protein from Clostridium thermocellum Cth-2968 | | Descriptor: | Putative translation initiation inhibitor, yjgF family, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Tempel, W, Lin, D, Zhang, H, Arendall III, W.B, Ljundahl, L, Liu, Z.-J, Rose, J, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-10-14 | | Release date: | 2004-12-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved hypothetical protein from Clostridium thermocellum Cth-2968
To be published
|
|
2B1M
 
 | | Crystal structure of a papain-fold protein without the catalytic cysteine from seeds of Pachyrhizus erosus | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPE31, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, M, Wei, Z, Chang, S. | | Deposit date: | 2005-09-16 | | Release date: | 2006-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a papain-fold protein without the catalytic residue: a novel member in the cysteine proteinase family
J.Mol.Biol., 358, 2006
|
|
3KCN
 
 | |
3KCM
 
 | |
3KXW
 
 | |
3L0Q
 
 | |
3OVG
 
 | | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound | | Descriptor: | PHOSPHATE ION, ZINC ION, amidohydrolase | | Authors: | Zhang, Z, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-16 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | The crystal structure of an amidohydrolase from Mycoplasma synoviae with Zn ion bound
To be Published
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
