7V3I
 
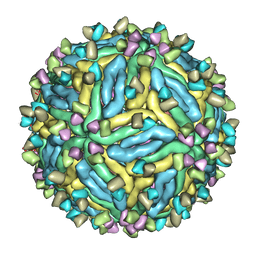 | | DENV2_NGC_Fab_C10 4degrees (3Fab:3E) | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
7V3J
 
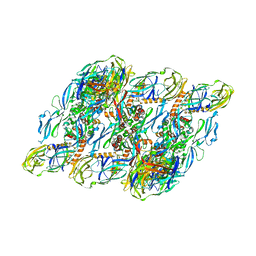 | | DENV2:F(ab')2-local | | Descriptor: | Envelope protein E, Fab_C10_heavy_chain, Fab_C10_light_chain, ... | | Authors: | Shu, B, Zhang, S, Victor, A.K, Ng, T.S, Lok, S.M. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Human antibody C10 neutralizes by diminishing Zika but enhancing dengue virus dynamics.
Cell, 184, 2021
|
|
3EBN
 
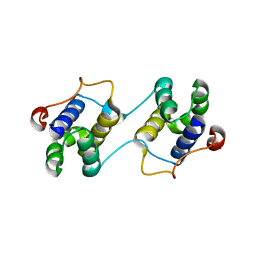 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | Deposit date: | 2008-08-28 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
3PKO
 
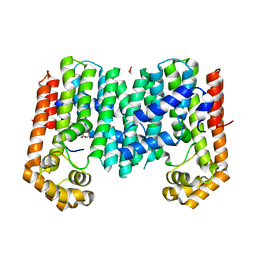 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 complexed with citrate | | Descriptor: | CITRIC ACID, GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4QFJ
 
 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Angiogenin, ... | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
7CN8
 
 | | Cryo-EM structure of PCoV_GX spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Wang, X, Yu, J, Zhang, S, Qiao, S, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
7CN4
 
 | | Cryo-EM structure of bat RaTG13 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Zhang, S, Qiao, S, Yu, J, Zeng, J, Tian, L. | | Deposit date: | 2020-07-30 | | Release date: | 2021-03-03 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Bat and pangolin coronavirus spike glycoprotein structures provide insights into SARS-CoV-2 evolution.
Nat Commun, 12, 2021
|
|
3I54
 
 | | Crystal structure of MtbCRP in complex with cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Transcriptional regulator, Crp/Fnr family | | Authors: | Reddy, M.C, Palaninathan, S.K, Bruning, J.B, Thurman, C, Smith, D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Mechanism of the Allosteric Transitions of Mycobacterium tuberculosis cAMP Receptor Protein.
J.Biol.Chem., 284, 2009
|
|
1NWA
 
 | | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine | | Descriptor: | Peptide methionine sulfoxide reductase msrA | | Authors: | Taylor, A.B, Benglis Jr, D.M, Dhandayuthapani, S, Hart, P.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-02-05 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Mycobacterium tuberculosis Methionine Sulfoxide Reductase A in Complex with Protein-bound Methionine
J.Bacteriol., 185, 2003
|
|
7X4B
 
 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7X2A
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | Descriptor: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
 | | S41 neutralizing antibody Fab(MERS-CoV) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.685 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
5DNK
 
 | | The structure of PKMT1 from Rickettsia prowazekii in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
5DPL
 
 | | The structure of PKMT2 from Rickettsia typhi in complex with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, protein lysine methyltransferase 2 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
2I6E
 
 | | Crystal structure of protein DR0370 from Deinococcus radiodurans, Pfam DUF178 | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Tyagi, R, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-28 | | Release date: | 2006-09-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of two proteins belonging to Pfam DUF178 revealed unexpected structural similarity to the DUF191 Pfam family.
Bmc Struct.Biol., 7, 2007
|
|
5H1S
 
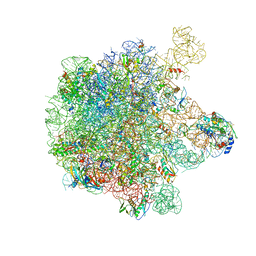 | | Structure of the large subunit of the chloro-ribosome | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L17, ... | | Authors: | Ahmed, T, Yin, Z, Bhushan, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-01 | | Last modified: | 2018-06-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the large subunit of the spinach chloroplast ribosome.
Sci Rep, 6, 2016
|
|
2QGO
 
 | |
4OHI
 
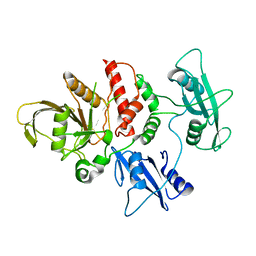 | | LEOPARD Syndrome-Associated SHP2/Q510E mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
2ISN
 
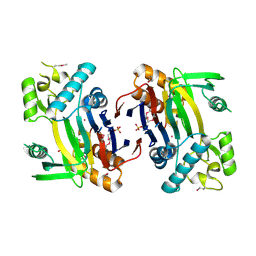 | | Crystal structure of a phosphatase from a pathogenic strain Toxoplasma gondii | | Descriptor: | NYSGXRC-8828z, phosphatase, PRASEODYMIUM ION, ... | | Authors: | Agarwal, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-18 | | Release date: | 2006-10-31 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
5ZEB
 
 | | M. Smegmatis P/P state 70S ribosome structure | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
7X28
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
3JQW
 
 | | Crystal structure of Clostridium histolyticum colH collagenase collagen-binding domain 3 at 2 Angstrom resolution in presence of calcium | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Sakon, J, Philominathan, S.T.L, Matsushita, O, Bauer, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Comparison of ColH and ColG Collagen-Binding Domains from Clostridium histolyticum.
J.Bacteriol., 195, 2013
|
|
3O4O
 
 | | Crystal structure of an Interleukin-1 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 beta, ... | | Authors: | Wang, X.Q, Wang, D.L, Zhang, S.Y, Li, L, Liu, X, Mei, K.R. | | Deposit date: | 2010-07-27 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the assembly and activation of IL-1beta with its receptors
Nat.Immunol., 11, 2010
|
|
5ZET
 
 | | M. smegmatis P/P state 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Mishra, S, Ahmed, T, Tyagi, A, Shi, J, Bhushan, S. | | Deposit date: | 2018-02-28 | | Release date: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of Mycobacterium smegmatis 70S ribosomes in complex with HPF, tmRNA, and P-tRNA.
Sci Rep, 8, 2018
|
|
