1ML9
 
 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | 分子名称: | Histone H3 methyltransferase DIM-5, UNKNOWN, ZINC ION | | 著者 | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | 登録日 | 2002-08-30 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
7X27
 
 | |
3L3S
 
 | |
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | 分子名称: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | 著者 | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | 登録日 | 2013-07-19 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
3T4W
 
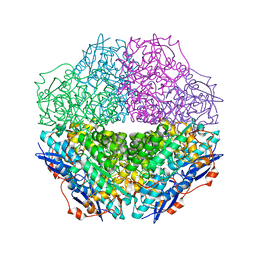 | | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp | | 分子名称: | Mandelate racemase/muconate lactonizing enzyme family protein | | 著者 | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2011-07-26 | | 公開日 | 2011-08-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | The crystal structure of mandelate racemase/muconate lactonizing enzyme from Sulfitobacter sp
To be Published
|
|
4DPO
 
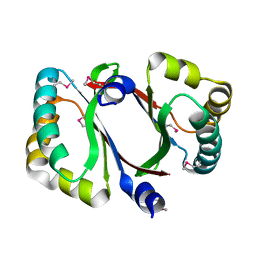 | | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1 | | 分子名称: | Conserved protein | | 著者 | Agarwal, R, Chamala, S, Evans, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Foti, R, Siedel, R, Zencheck, W, Villigas, G, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-13 | | 公開日 | 2012-02-29 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Crystal structure of a conserved protein MM_1583 from Methanosarcina mazei Go1
To be Published
|
|
4DQX
 
 | | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42 | | 分子名称: | Probable oxidoreductase protein | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-16 | | 公開日 | 2012-02-29 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of a short chain dehydrogenase from Rhizobium etli CFN 42
To be Published
|
|
3KOL
 
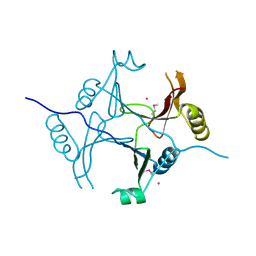 | |
7X2A
 
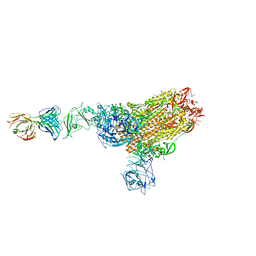 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class1 (1u2d RBD with 1Fab) | | 分子名称: | MERS-CoV Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X26
 
 | | S41 neutralizing antibody Fab(MERS-CoV) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2022-11-23 | | 実験手法 | ELECTRON MICROSCOPY (3.685 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
7X29
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class2 (1u2d RBD with 2Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
1MQ7
 
 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | 著者 | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2002-09-13 | | 公開日 | 2002-10-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
7X28
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class3 (2u1d RBD with 2Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J.W, Zhang, S.Y, Zhou, H.X, Wang, X.W. | | 登録日 | 2022-02-25 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
4DYV
 
 | | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2 | | 分子名称: | CHLORIDE ION, Short-chain dehydrogenase/reductase SDR | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Gizzi, A, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-02-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a short-chain dehydrogenase/reductase SDR from Xanthobacter autotrophicus Py2
To be Published
|
|
4E1J
 
 | | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021 | | 分子名称: | CHLORIDE ION, GLYCEROL, Glycerol kinase, ... | | 著者 | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Siedel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-06 | | 公開日 | 2012-03-21 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of glycerol kinase in complex with glycerol from Sinorhizobium meliloti 1021
To be Published
|
|
4E3Z
 
 | | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42 | | 分子名称: | Putative oxidoreductase protein | | 著者 | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-03-11 | | 公開日 | 2012-03-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of a oxidoreductase from Rhizobium etli CFN 42
To be Published
|
|
2QGO
 
 | |
2G59
 
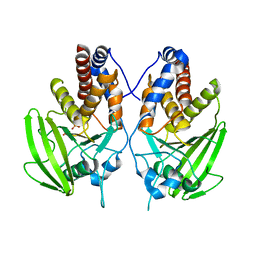 | |
3KTN
 
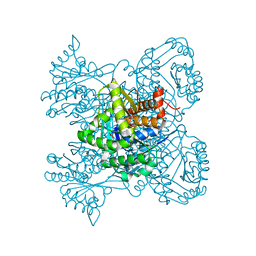 | |
2QS8
 
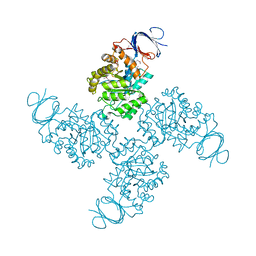 | | Crystal structure of a Xaa-Pro dipeptidase with bound methionine in the active site | | 分子名称: | MAGNESIUM ION, METHIONINE, Xaa-Pro Dipeptidase | | 著者 | Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-07-30 | | 公開日 | 2007-08-21 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Functional annotation of two new carboxypeptidases from the amidohydrolase superfamily of enzymes.
Biochemistry, 48, 2009
|
|
8J67
 
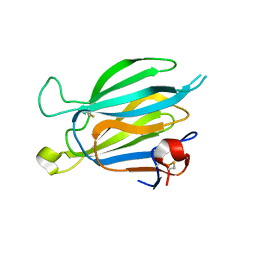 | | Crystal structure of Toxoplasma gondii M2AP | | 分子名称: | MIC2-associated protein | | 著者 | Wang, F.F, Zhang, D.J, Zhang, S, Springer, T.A, Song, G.J. | | 登録日 | 2023-04-24 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural insights into MIC2 recognition by MIC2-associated protein in Toxoplasma gondii.
Commun Biol, 6, 2023
|
|
3KZH
 
 | |
7X25
 
 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | 分子名称: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | 著者 | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | 登録日 | 2022-02-25 | | 公開日 | 2023-01-18 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (2.49 Å) | | 主引用文献 | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | 分子名称: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-06-25 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
3KTO
 
 | |
