8HNB
 
 | | Cryo-EM structure of human OATP1B1 in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1 | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNH
 
 | | Cryo-EM structure of human OATP1B1 in complex with simeprevir | | Descriptor: | (2R,3aR,10Z,11aS,12aR,14aR)-N-(cyclopropylsulfonyl)-2-({7-methoxy-8-methyl-2-[4-(1-methylethyl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-4,14-dioxo-2,3,3a,4,5,6,7,8,9,11a,12,13,14,14a-tetradecahydrocyclopenta[c]cyclopropa[g][1,6]diazacyclotetradecine-12a(1H)-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shan, Z, Yang, X, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
3O5X
 
 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
5JO4
 
 | | Antibody Fab Fragment Complex | | Descriptor: | D80 Fab Heavy Chain, D80 Fab Light Chain, G6 Fab Heavy Chain, ... | | Authors: | Zhang, Z, Prachanronarong, K.P, Marasco, W.A, Schiffer, C.A.S. | | Deposit date: | 2016-05-01 | | Release date: | 2017-11-08 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
5JQD
 
 | | Antibody Fab Fragment | | Descriptor: | D80 Fab Fragment Heavy Chain, D80 Fab Fragment Light Chain | | Authors: | Zhang, Z, Prachanronarong, K, Gellatly, K, Marasco, W.A, Schiffer, C.A. | | Deposit date: | 2016-05-04 | | Release date: | 2017-11-08 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
6Y6I
 
 | | lipopolysaccharide outer core galactosyltransferase WaaB and UDP complex | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
6Y6G
 
 | | lipopolysaccharide outer core galactosyltransferase WaaB apo form | | Descriptor: | Lipopolysaccharide 1,6-galactosyltransferase | | Authors: | Zhang, Z.Y, Ashworth, G, Wang, Z.S, Zhu, X.F, Dong, C.J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into the lipopolysaccharide outer core galactosyltransferase WaaB
To Be Published
|
|
1GVG
 
 | | Crystal Structure of Clavaminate Synthase with Nitric Oxide | | Descriptor: | 2-OXOGLUTARIC ACID, CLAVAMINATE SYNTHASE 1, DEOXYGUANIDINOPROCLAVAMINIC ACID, ... | | Authors: | Zhang, Z.H, Ren, J, McKinnon, C.H, Clifton, I.J, Harlos, K, Schofield, C.J. | | Deposit date: | 2002-02-12 | | Release date: | 2003-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of a Clavaminate Synthase-Fe(II) -2-Oxoglutarate-Substrate-No Complex: Evidence for Metal Centered Rearrangements
FEBS Lett., 517, 2002
|
|
1HQW
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CONCANAVALIN A WITH A TRIPEPTIDE YPY | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y, Wang, K, Cui, D, Li, M. | | Deposit date: | 2000-12-20 | | Release date: | 2003-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of concanavalin A and tripeptide
J.PROTEIN CHEM., 20, 2001
|
|
3HDC
 
 | |
8GUN
 
 | |
8GUO
 
 | |
6Y5Q
 
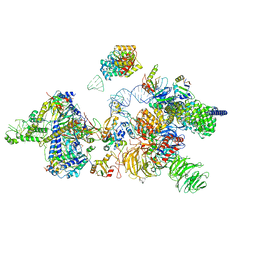 | | human 17S U2 snRNP | | Descriptor: | HIV Tat-specific factor 1, PHD finger-like domain-containing protein 5A, Probable ATP-dependent RNA helicase DDX46, ... | | Authors: | Zhang, Z, Will, C.L, Bertram, K, Luehrmann, R, Stark, H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular architecture of the human 17S U2 snRNP.
Nature, 583, 2020
|
|
3FKD
 
 | |
8GXN
 
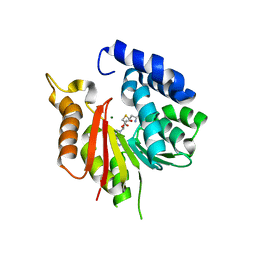 | | The crystal structure of CsFAOMT2 in complex with SAH | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, caffeyl-CoA-O-methyltransferase | | Authors: | Zhang, Z.M, Zhou, Y.E, Huang, H.S. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Characterization of two O-methyltransferases involved in the biosynthesis of O-methylated catechins in tea plant.
Nat Commun, 14, 2023
|
|
8GXO
 
 | |
3EVZ
 
 | |
6F57
 
 | |
7OQE
 
 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
8BDP
 
 | |
3MSR
 
 | |
3GG4
 
 | |
3GBT
 
 | |
3DZB
 
 | |
