8WHB
 
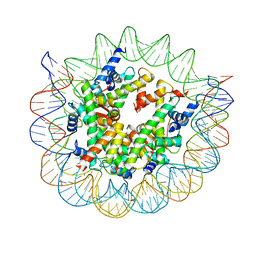 | | Structure of nucleosome core particle of Arabidopsis thaliana | | Descriptor: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH5
 
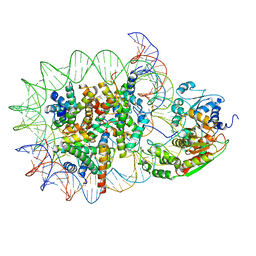 | | Structure of DDM1-nucleosome complex in the apo state | | Descriptor: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | Authors: | Liu, Y, Zhang, Z, Du, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8XV9
 
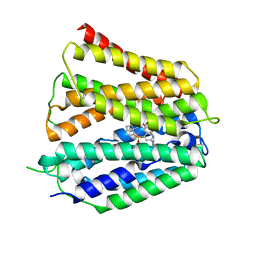 | | Fedratinib-bound human SLC19A3 | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
1EEO
 
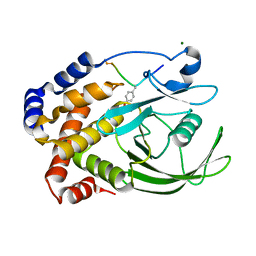 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
4UI9
 
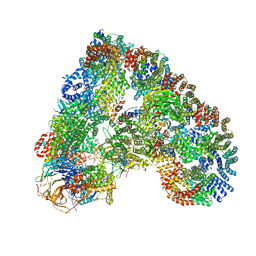 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
8WX3
 
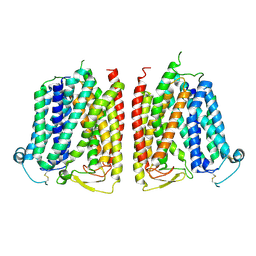 | |
8WX2
 
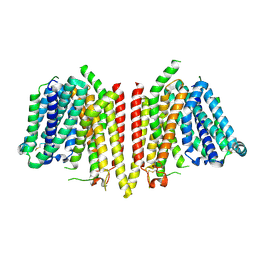 | |
8WX4
 
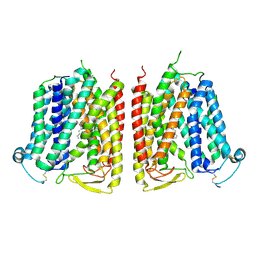 | |
8WX5
 
 | |
5W3T
 
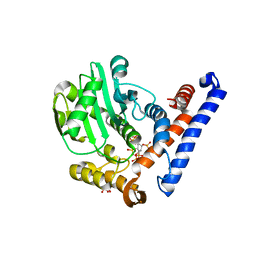 | | Crystal structure of PopP2 in complex with IP6 | | Descriptor: | GLYCEROL, INOSITOL HEXAKISPHOSPHATE, PopP2 protein | | Authors: | Song, J, Zhang, Z.M. | | Deposit date: | 2017-06-08 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of host substrate acetylation by a YopJ family effector.
Nat Plants, 3, 2017
|
|
8XV2
 
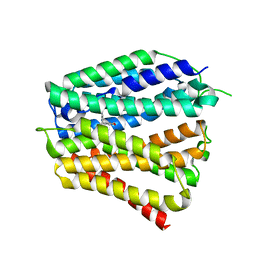 | | Thiamine-bound human SLC19A3 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
8XV5
 
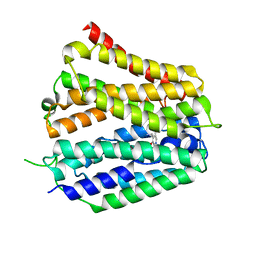 | | Pyridoxamine-bound human SLC19A3 | | Descriptor: | 4-(AMINOMETHYL)-5-(HYDROXYMETHYL)-2-METHYLPYRIDIN-3-OL, Soluble cytochrome b562,Thiamine transporter 2 | | Authors: | Dang, Y, Wang, G.P, Zhang, Z. | | Deposit date: | 2024-01-14 | | Release date: | 2024-03-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate and drug recognition mechanisms of SLC19A3.
Cell Res., 34, 2024
|
|
8IDN
 
 | | Cryo-EM structure of RBD/E77-Fab complex | | Descriptor: | E77 Fab heavy chain, E77 Fab light chain, Spike protein S1, ... | | Authors: | Lu, D.F, Zhang, Z.C. | | Deposit date: | 2023-02-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | The structure of the RBD-E77 Fab complex reveals neutralization and immune escape of SARS-CoV-2.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7YVB
 
 | | Aplysia californica FaNaC in ligand bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated Na+ channel, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
7YVC
 
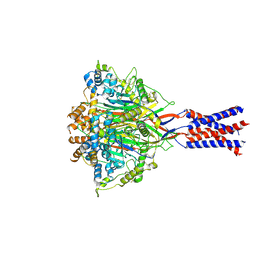 | | Aplysia californica FaNaC in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
8A5Y
 
 | | S. cerevisiae apo unphosphorylated APC/C. | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex and comparison to apo unphosphorylated and phosphorylated states
To Be Published
|
|
8A61
 
 | | S. cerevisiae apo phosphorylated APC/C | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Fernandez-Vazquez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae apo phosphorylated APC/C
To Be Published
|
|
8A3T
 
 | | S. cerevisiae APC/C-Cdh1 complex | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 11, Anaphase-promoting complex subunit 2, ... | | Authors: | Barford, D, Vazquez-Fernandez, E, Zhang, Z, Yang, J. | | Deposit date: | 2022-06-09 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the S. cerevisiae APC/C-Cdh1 complex
To Be Published
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7D3Y
 
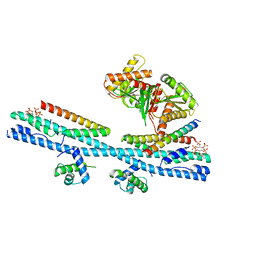 | | Crystal structure of the osPHR2-osSPX2 complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Protein PHOSPHATE STARVATION RESPONSE 2, SPX domain-containing protein 2,Isoform 1 of Core histone macro-H2A.1 | | Authors: | Zhang, Q.X, Guan, Z.Y, Zuo, J.Q, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
6LYH
 
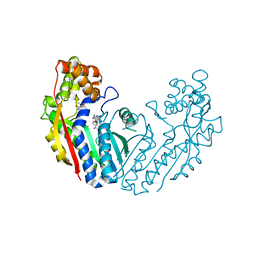 | | Crystal structure of tea N9-methyltransferase CkTcS in complex with SAH and 1,3,7-trimethyluric acid | | Descriptor: | 1,3,7-trimethyl-9H-purine-2,6,8-trione, N-methyltransferase CkTcS, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wang, Y, Zhang, Z.-M. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.15000319 Å) | | Cite: | Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun, 11, 2020
|
|
7D3T
 
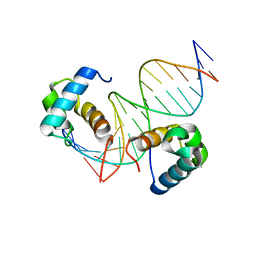 | | Crystal structure of OSPHR2 in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*TP*CP*GP*GP*AP*TP*AP*TP*CP*CP*TP*CP*AP*AP*G)-3'), DNA (5'-D(P*GP*CP*TP*TP*GP*AP*GP*GP*AP*TP*AP*TP*CP*CP*GP*A)-3'), Protein PHOSPHATE STARVATION RESPONSE 2 | | Authors: | Guan, Z.Y, Zhang, Z.F, Liu, Z. | | Deposit date: | 2020-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2 - PHR2 complex.
Nat Commun, 13, 2022
|
|
6KZD
 
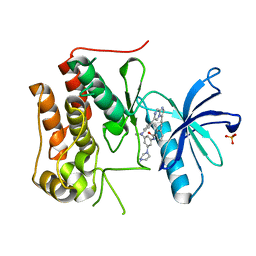 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
6M4Q
 
 | | Cytochrome P450 monooxygenase StvP2 substrate-free structure | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
