7X0X
 
 | |
7X0Y
 
 | |
8KC7
 
 | | Rpd3S histone deacetylase complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Dong, S, Li, H, Wang, M, Rasheed, N, Zou, B, Gao, X, Guan, J, Li, W, Zhang, J, Wang, C, Zhou, N, Shi, X, Li, M, Zhou, M, Huang, J, Li, H, Zhang, Y, Wong, K.H, Zhang, X, Chao, W.C.H, He, J. | | Deposit date: | 2023-08-06 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of nucleosome deacetylation and DNA linker tightening by Rpd3S histone deacetylase complex.
Cell Res., 33, 2023
|
|
7QTQ
 
 | | Structure of Native, iodinated bovine thyroglobulin solved on strepavidin affinity grids. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thyroglobulin, ... | | Authors: | Marechal, N, Weitz, J.C, Serrano, B.P, Zhang, X. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Formation of thyroid hormone revealed by a cryo-EM structure of native bovine thyroglobulin.
Nat Commun, 13, 2022
|
|
7QXI
 
 | |
7QV9
 
 | |
7QWP
 
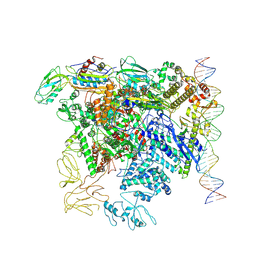 | | CryoEM structure of bacterial transcription close complex (RPc) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Ye, F.Z, Zhang, X.D. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanisms of DNA opening revealed in AAA+ transcription complex structures.
Sci Adv, 8, 2022
|
|
7C4J
 
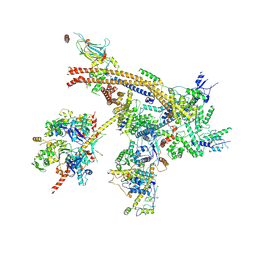 | | Cryo-EM structure of the yeast Swi/Snf complex in a nucleosome free state | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Regulator of Ty1 transposition protein 102, ... | | Authors: | Wang, C.C, Guo, Z.Y, Zhan, X.C, Zhang, X.F. | | Deposit date: | 2020-05-18 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the yeast Swi/Snf complex in a nucleosome free state
Nat Commun, 2020
|
|
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
3ZN8
 
 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
8JEQ
 
 | | Crystal structure of Tiragolumab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEP
 
 | | Crystal structure of Ociperlimab | | Descriptor: | antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
8JEO
 
 | | Crystal structure of TIGIT in complexed with Tiragolumab | | Descriptor: | T-cell immunoreceptor with Ig and ITIM domains, antibody heavy chain, antibody light chain | | Authors: | Sun, J, Zhang, X.X, Song, J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural insights into the unique pH-responsive characteristics of the anti-TIGIT therapeutic antibody Ociperlimab.
Structure, 32, 2024
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJW
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-phenylazanylbenzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7X5A
 
 | | CryoEM structure of RuvA-Holliday junction complex | | Descriptor: | DNA (26-MER), Holliday junction ATP-dependent DNA helicase RuvA | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7P
 
 | | CryoEM structure of dsDNA-RuvB-RuvA domain3 complex | | Descriptor: | DNA, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
7X7Q
 
 | | CryoEM structure of RuvA-RuvB-Holliday junction complex | | Descriptor: | DNA (26-MER), DNA (40-MER), Holliday junction ATP-dependent DNA helicase RuvA, ... | | Authors: | Lin, Z, Qu, Q, Zhang, X, Zhou, Z. | | Deposit date: | 2022-03-10 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Cryo-EM structure of the RuvAB-Holliday junction intermediate complex from Pseudomonas aeruginosa.
Front Plant Sci, 14, 2023
|
|
8Y8I
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-3up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8E
 
 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
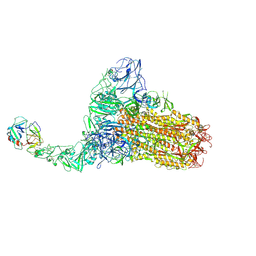 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
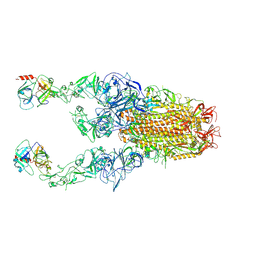 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8A
 
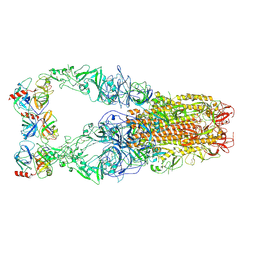 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
