4RVU
 
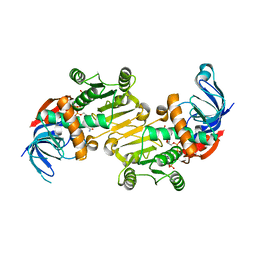 | | The native structure of mycobacterial Rv1454c complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable quinone reductase Qor (NADPH:quinone reductase) (Zeta-crystallin homolog protein) | | Authors: | Zhou, W.H, Zheng, Q.Q, Song, Y.L, Zhang, W, Shaw, N, Rao, Z. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7988 Å) | | Cite: | Structural views of quinone oxidoreductase from Mycobacterium tuberculosis reveal large conformational changes induced by the co-factor.
Febs J., 282, 2015
|
|
6RV4
 
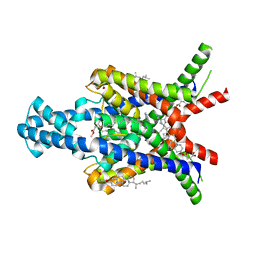 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6GX5
 
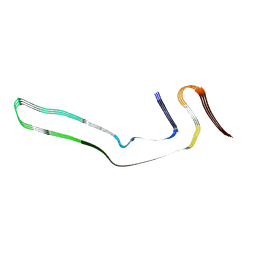 | | Narrow Pick Filament from Pick's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Murzin, A.G, Murshudov, G, Garringer, H.J, Vidal, R, Crowther, R.A, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-06-26 | | Release date: | 2018-09-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of filaments from Pick's disease reveal a novel tau protein fold.
Nature, 561, 2018
|
|
6RV3
 
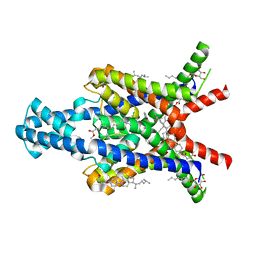 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 1000493 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
5JG6
 
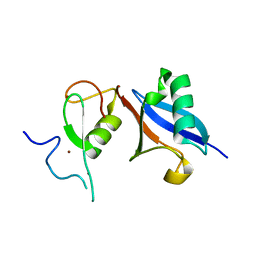 | | APC11-Ubv shows role of noncovalent RING-Ubiquitin interactions in processive multiubiquitination and Ubiquitin chain elongation by APC/C | | Descriptor: | Anaphase-promoting complex subunit 11, Polyubiquitin-B, ZINC ION | | Authors: | Brown, N.G, Zhang, W, Yu, S, Miller, D.J, Sidhu, S.S, Schulman, B.A. | | Deposit date: | 2016-04-19 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0013 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
7MOO
 
 | | L-type DNA containing 2'-fluoro-2'-deoxycytidine | | Descriptor: | DNA (5'-D(*(0DG)P*(OFC)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3') | | Authors: | Dantsu, Y, Zhang, Y, Zhang, W. | | Deposit date: | 2021-05-03 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis of 2'-Deoxy-2'-fluoro-L-cytidine and Fluorinated L-Nucleic Acids for Structural Studies
Chemistryselect, 6, 2021
|
|
5K0K
 
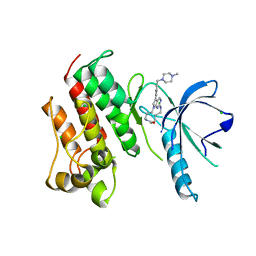 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2434 | | Descriptor: | 15-{4-[(4-methylpiperazin-1-yl)methyl]phenyl}-4,5,6,7,9,10,11,12-octahydro-2,16-(azenometheno)pyrrolo[2,1-d][1,3,5,9]te traazacyclotetradecin-8(3H)-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, X, Liu, J, Zhang, W, Stashko, M.A, Nichols, J, DeRyckere, D, Miley, M.J, Norris-Drouin, J, Chen, Z, Machius, M, Wood, E, Graham, D.K, Earp, H.S, Graham, K, Kireev, D, Frye, S.V. | | Deposit date: | 2016-05-17 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Design and Synthesis of Novel Macrocyclic Mer Tyrosine Kinase Inhibitors.
ACS Med Chem Lett, 7, 2016
|
|
5K0X
 
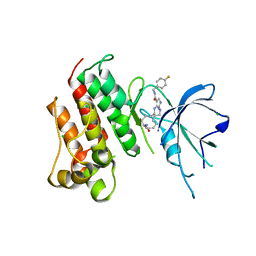 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC2541 | | Descriptor: | (7S)-7-amino-N-[(4-fluorophenyl)methyl]-8-oxo-2,9,16,18,21-pentaazabicyclo[15.3.1]henicosa-1(21),17,19-triene-20-carboxamide, CHLORIDE ION, Tyrosine-protein kinase Mer | | Authors: | McIver, A.L, Zhang, W, Liu, Q, Jiang, X, Stashko, M.A, Nichols, J, Miley, M.J, Norris-Drouin, J, Machius, M, DeRyckere, D, Wood, E, Graham, D.K, Earp, H.S, Kireev, D, Frye, S.V, Wang, X. | | Deposit date: | 2016-05-17 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Discovery of Macrocyclic Pyrimidines as MerTK-Specific Inhibitors.
ChemMedChem, 12, 2017
|
|
1RWT
 
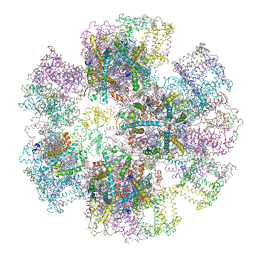 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
6CV0
 
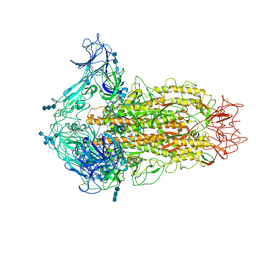 | | Cryo-electron microscopy structure of infectious bronchitis coronavirus spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Luo, C, Zhang, W, Li, F. | | Deposit date: | 2018-03-27 | | Release date: | 2018-04-18 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins.
PLoS Pathog., 14, 2018
|
|
6OWL
 
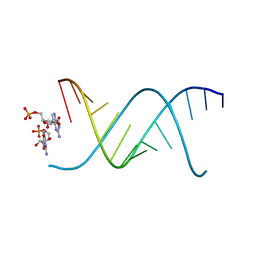 | | RNA oligonucleotides with 3'-arabino guanosine co-crystallized with GMP | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*R)-3'), RNA (5'-R(P*G)-3') | | Authors: | Szostak, J.W, Kim, S, Zhang, W. | | Deposit date: | 2019-05-10 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Model for the Emergence of RNA from a Prebiotically Plausible Mixture of Ribonucleotides, Arabinonucleotides, and 2'-Deoxynucleotides.
J.Am.Chem.Soc., 142, 2020
|
|
4V52
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3RTP
 
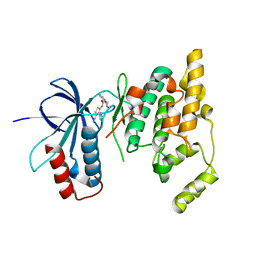 | | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration | | Descriptor: | Mitogen-activated protein kinase 10, N-[4-cyano-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, R.J, Hom, R.K, Sealy, J.M, Probst, G.D, Quincy, Q, Peterson, B, Chan, W, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Pan, H, Lin, M, Yao, N, Artis, D.R, Zhang, H, Chen, L, Dryer, M, Samant, B, Zmolek, W, Wong, K, Lorentzen, C, Goldbach, E, Tonn, G, Quinn, K.P, Sauer, J, Wright, S, Powell, K, Ruslim, L, Ren, Z, Bard, F, Yednock, T.A, Griswold-Prenne, I. | | Deposit date: | 2011-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6U9I
 
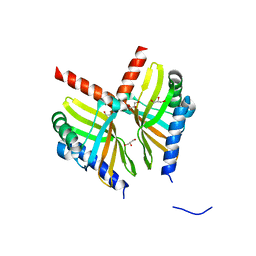 | | Crystal structure of BvnE pinacolase from Penicillium brevicompactum | | Descriptor: | BvnE, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Ye, Y, Du, L, Zhang, X, Newmister, S.A, McCauley, M, Alegre-Requena, J.V, Zhang, W, Mu, S, Minami, A, Fraley, A.E, Adrover-Castellano, M.L, Carney, N, Shende, V.V, Oikawa, H, Kato, H, Tsukamoto, S, Paton, R.S, Williams, R.M, Sherman, D.H, Li, S. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | Fungal-derived brevianamide assembly by a stereoselective semipinacolase.
Nat Catal, 3, 2020
|
|
3VAY
 
 | | Crystal structure of 2-Haloacid Dehalogenase from Pseudomonas syringae pv. Tomato DC3000 | | Descriptor: | HAD-superfamily hydrolase, IODIDE ION, MAGNESIUM ION | | Authors: | Hou, Z, Zhang, H, Li, M, Chang, W. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 2-haloacid dehalogenase from Pseudomonas syringae pv. tomato DC3000
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8OR1
 
 | | Co-crystal strucutre of PD-L1 with low molecular weight inhibitor | | Descriptor: | 5-[[5-[[2-chloranyl-3-(2-fluorophenyl)phenyl]methoxy]-2-[(~{E})-2-hydroxyethyliminomethyl]phenoxy]methyl]pyridine-3-carbonitrile, Programmed cell death 1 ligand 1 | | Authors: | Zhang, H, Zhou, S, Wu, C, Zhu, M, Yu, Q, Wang, X, Awadasseid, A, Plewka, J, Magiera-Mularz, K, Wu, Y, Zhang, W. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, Synthesis, and Antitumor Activity Evaluation of 2-Arylmethoxy-4-(2,2'-dihalogen-substituted biphenyl-3-ylmethoxy) Benzylamine Derivatives as Potent PD-1/PD-L1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
7N7X
 
 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
8F24
 
 | | Mirror-image RNA octamer containing 2'-OMe-L-uridine | | Descriptor: | Mirror-image RNA 0G-XEC-0G-0U-0A-0C-0A-0C, SULFATE ION | | Authors: | Dantsu, Y, Zhang, W. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
8FZC
 
 | |
5MDN
 
 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNB
 
 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNA
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8CMZ
 
 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
