3FVS
 
 | | Human Kynurenine Aminotransferase I in complex with Glycerol | | Descriptor: | GLYCEROL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3DHE
 
 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED DEHYDROEPIANDROSTERONE | | Descriptor: | 3-BETA-HYDROXY-5-ANDROSTEN-17-ONE, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Han, Q, Campbell, R.L, Gangloff, A, Lin, S.X. | | Deposit date: | 1998-03-25 | | Release date: | 1999-09-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
J.Biol.Chem., 275, 2000
|
|
3DC1
 
 | | Crystal structure of kynurenine aminotransferase II complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, GLYCEROL, Kynurenine/alpha-aminoadipate aminotransferase mitochondrial | | Authors: | Han, Q, Cai, T, Tagle, D.A, Robinson, H, Li, J. | | Deposit date: | 2008-06-03 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate specificity and structure of human aminoadipate aminotransferase/kynurenine aminotransferase II
Biosci.Rep., 28, 2008
|
|
3FVU
 
 | | Crystal Structure of Human Kynurenine Aminotransferase I in Complex with Indole-3-acetic Acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, GLYCEROL, Kynurenine--oxoglutarate transaminase 1, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
3FVX
 
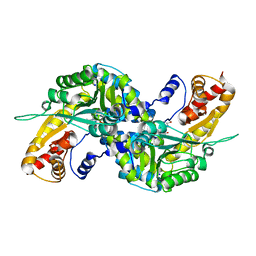 | | Human kynurenine aminotransferase I in complex with tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Kynurenine--oxoglutarate transaminase 1, SODIUM ION | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
2QLR
 
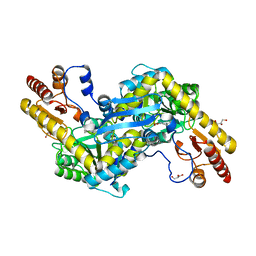 | |
7JMU
 
 | | Hen egg-white lysozyme with ionic liquid ethylammonium nitrate | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Han, Q, Darmanin, C, Smith, K, Greaves, T, Drummond, C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Lysozyme conformational changes with ionic liquids: Spectroscopic, small angle x-ray scattering and crystallographic study.
J Colloid Interface Sci, 585, 2021
|
|
1DHT
 
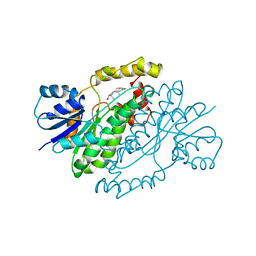 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED DIHYDROTESTOSTERONE | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Han, Q, Campbell, R.L, Gangloff, A, Lin, S.X. | | Deposit date: | 1998-03-25 | | Release date: | 1999-09-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Dehydroepiandrosterone and dihydrotestosterone recognition by human estrogenic 17beta-hydroxysteroid dehydrogenase. C-18/c-19 steroid discrimination and enzyme-induced strain.
J.Biol.Chem., 275, 2000
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
6V3X
 
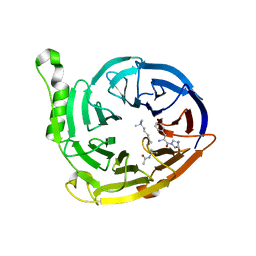 | |
6V3Y
 
 | |
9JRI
 
 | | outward-open hSLC19A1 + 5-MTHF | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRK
 
 | | outward-open hSLC19A1 + MTX | | Descriptor: | METHOTREXATE, Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRL
 
 | | outward-open hSLC19A1 + PT523 | | Descriptor: | N-(4-CARBOXY-4-{4-[(2,4-DIAMINO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-BUTYL)-PHTHALAMIC ACID, Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JOZ
 
 | | outward-open hSLC19A1 | | Descriptor: | Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-25 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRN
 
 | | outward-open hSLC19A1 + TPP | | Descriptor: | Soluble cytochrome b562,Reduced folate transporter,fusion protein, THIAMINE DIPHOSPHATE | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
9JRM
 
 | | outward-open hSLC19A1 + 2'3'-CDAS | | Descriptor: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Soluble cytochrome b562,Reduced folate transporter,Soluble cytochrome b562,Reduced folate transporter,fusion protein | | Authors: | Zhang, Q.X, Liu, K.X, Gao, P. | | Deposit date: | 2024-09-29 | | Release date: | 2025-02-26 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport.
Nat Commun, 16, 2025
|
|
8GOF
 
 | | Structure of hSLC19A1+PMX | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
5DV2
 
 | |
8UKD
 
 | |
8UIR
 
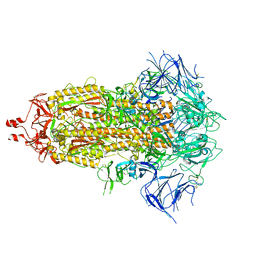 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8UKF
 
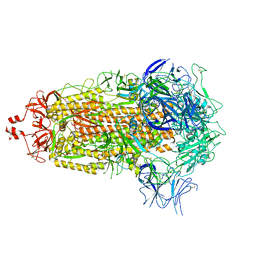 | | SARS-CoV-2 Omicron-EG.5 3-RBD down Spike Protein Trimer consensus (S-GSAS-Omicron-EG.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
8UK1
 
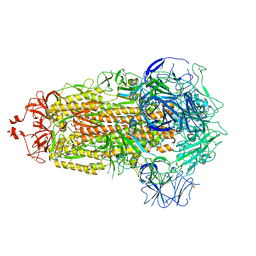 | | SARS-CoV-2 Omicron-XBB.1.16 3-RBD-down Spike Protein Trimer consensus (S-RRAR-Omicron-XBB.1.16) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, Q.E, Acharya, P. | | Deposit date: | 2023-10-11 | | Release date: | 2024-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SARS-CoV-2 Omicron XBB lineage spike structures, conformations, antigenicity, and receptor recognition.
Mol.Cell, 84, 2024
|
|
