8GOE
 
 | | Structure of hSLC19A1+5-MTHF | | Descriptor: | N-[4-({[(6S)-2-AMINO-4-HYDROXY-5-METHYL-5,6,7,8-TETRAHYDROPTERIDIN-6-YL]METHYL}AMINO)BENZOYL]-L-GLUTAMIC ACID, Reduced folate transporter | | Authors: | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | Deposit date: | 2022-08-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
8GV9
 
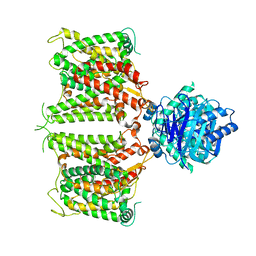 | | The cryo-EM structure of hAE2 with chloride ion | | Descriptor: | Anion exchange protein 2, CHLORIDE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
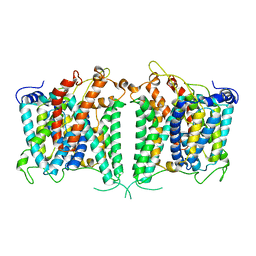 | | The intermediate structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
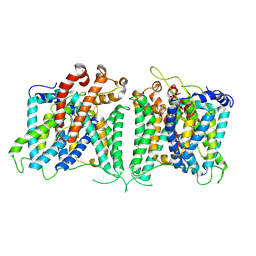 | | The asymmetry structure of hAE2 | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
1PQQ
 
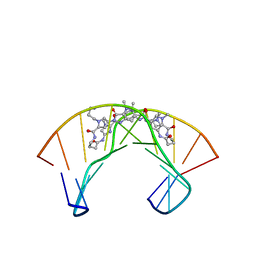 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
8GVC
 
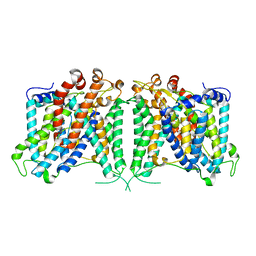 | | The cryo-EM structure of hAE2 with bicarbonate | | Descriptor: | Anion exchange protein 2, BICARBONATE ION | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
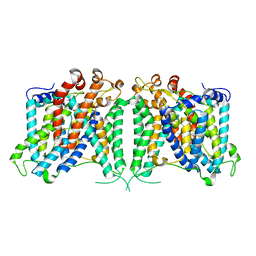 | | The outward-facing structure of hAE2 in basic pH | | Descriptor: | Anion exchange protein 2 | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
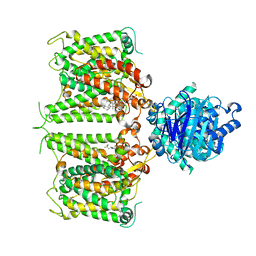 | | Human AE2 in acidic KNO3 | | Descriptor: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | Authors: | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
5DV2
 
 | |
5DV4
 
 | |
6V3X
 
 | |
6V3Y
 
 | |
2DO8
 
 | | Solution Structure of UPF0301 protein HD_1794 | | Descriptor: | UPF0301 protein HD_1794 | | Authors: | Zhang, Q, Liu, G, Shastry, R, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T.R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-27 | | Release date: | 2006-05-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of UPF0301 protein HD_1794
To be published
|
|
5KH8
 
 | |
2H3J
 
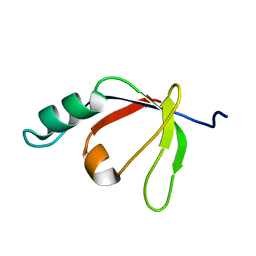 | | Solution NMR Structure of Protein PA4359 from Pseudomonas aeruginosa: Northeast Structural Genomics Consortium Target PaT89 | | Descriptor: | Hypothetical protein PA4359 | | Authors: | Zhang, Q, Liu, G, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-22 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Hypothetical protein PA4359: Northest Structural Genomics Target PaT89
To be Published
|
|
2L3E
 
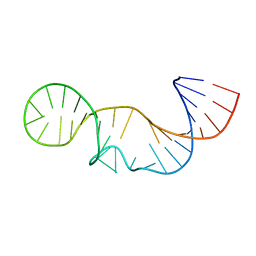 | | Solution structure of P2a-J2a/b-P2b of human telomerase RNA | | Descriptor: | 35-MER | | Authors: | Zhang, Q, Kim, N, Peterson, R.D, Wang, Z, Feigon, J. | | Deposit date: | 2010-09-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inaugural Article: Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
8B2X
 
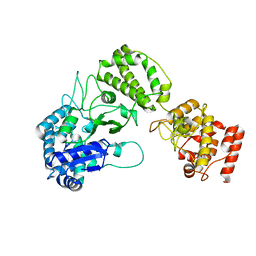 | |
8ANE
 
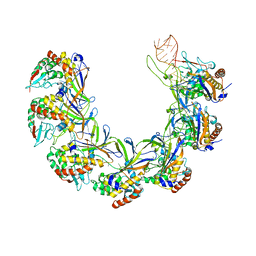 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
4JGV
 
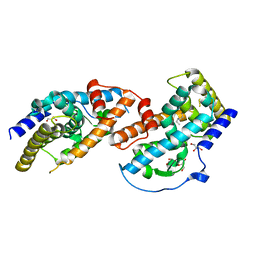 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
5YJZ
 
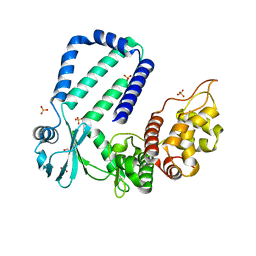 | | The native crystal structure of Rv3197 from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, Probable conserved ATP-binding protein ABC transporter, SULFATE ION | | Authors: | Zhang, Q.Q, Rao, Z.H. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of the first macrolide antibiotic binding protein in Mycobacterium tuberculosis: a new antibiotic resistance drug target.
Protein Cell, 9, 2018
|
|
2PEE
 
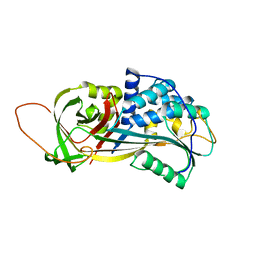 | | Crystal Structure of a Thermophilic Serpin, Tengpin, in the Native State | | Descriptor: | GLYCEROL, SULFATE ION, Serine protease inhibitor | | Authors: | Zhang, Q.W, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2007-04-02 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N terminus of the serpin, tengpin, functions to trap the metastable native state.
Embo Rep., 8, 2007
|
|
