3QG1
 
 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3TNQ
 
 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein kinase, ... | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3TNP
 
 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3UVM
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | Descriptor: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3URO
 
 | | Poliovirus receptor CD155 D1D2 | | Descriptor: | Poliovirus receptor | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5005 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3UVO
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1B | | Descriptor: | Histone-lysine N-methyltransferase SETD1B, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVN
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | Descriptor: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVL
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL3 | | Descriptor: | Histone-lysine N-methyltransferase MLL3, WD repeat-containing protein 5 | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVK
 
 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase MLL2, SULFATE ION, ... | | Authors: | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3HMJ
 
 | | Saccharomyces cerevisiae FAS type I | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Johansson, P, Mulinacci, B, Koestler, C, Vollrath, R, Oesterhelt, D, Grininger, M. | | Deposit date: | 2009-05-29 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Structure, 17, 2009
|
|
4PO2
 
 | | Crystal Structure of the Stress-Inducible Human Heat Shock Protein HSP70 Substrate-Binding Domain in Complex with Peptide Substrate | | Descriptor: | HSP70 substrate peptide, Heat shock 70 kDa protein 1A/1B, PHOSPHATE ION, ... | | Authors: | Zhang, P, Leu, J.I, Murphy, M.E, George, D.L, Marmorstein, R. | | Deposit date: | 2014-02-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the stress-inducible human heat shock protein 70 substrate-binding domain in complex with Peptide substrate.
Plos One, 9, 2014
|
|
1JDG
 
 | | Solution Structure of a Trans-Opened (10S)-dA Adduct of (+)-(7S,8R,9S,10R)-7,8-Dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully Complementary DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*GP*TP*GP*AP*CP*CP*G)-3', 5'-D(*CP*GP*GP*TP*CP*(BPA)AP*CP*GP*AP*GP*G)-3', 7S,8R,9R-TRIHYDROXY-7,8,9,10-TETRAHYDRO BENZO[A]PYRENE | | Authors: | Pradhan, P, Tirumala, S, Liu, X, Sayer, J.M, Jerina, D.M, Yeh, H.J.C. | | Deposit date: | 2001-06-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a trans-opened (10S)-dA adduct of (+)-(7S,8R,9S,10R)-7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene in a fully complementary DNA duplex: evidence for a major syn conformation.
Biochemistry, 40, 2001
|
|
2WAS
 
 | |
3J82
 
 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
7V1A
 
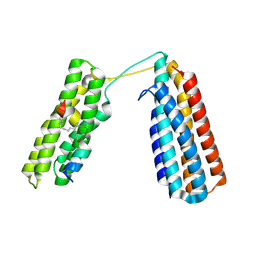 | | Stapled TBS peptide from RIAM bound to talin R7R8 domains | | Descriptor: | 1,2-ETHANEDIOL, ASP-ILE-ASP-GLN-MET-PHE-SER-THR-LEU-LEU-GLY-GLU-MK8-ASP-LEU-LEU-MK8-GLN-SER, Talin-1 | | Authors: | Zhang, P, Gao, T, Wu, J. | | Deposit date: | 2022-05-11 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibition of talin-induced integrin activation by a double-hit stapled peptide.
Structure, 31, 2023
|
|
4AJF
 
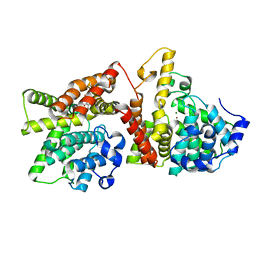 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJD
 
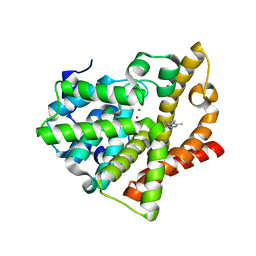 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | 2-ETHYL-4-METHYL-PHTHALAZIN-1-ONE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
4AJG
 
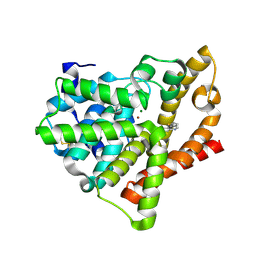 | | Identification and structural characterization of PDE10 fragment inhibitors | | Descriptor: | CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, MAGNESIUM ION, ... | | Authors: | Johansson, P, Albert, J.S, Spadola, L, Akerud, T, Back, E, Hillertz, P, Horsefeld, R, Scott, C, Spear, N, Tian, G, Tigerstrom, A, Aharony, D, Geschwindner, S. | | Deposit date: | 2012-02-16 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Structural Characterization of Pde10 Fragment Inhibitors
To be Published
|
|
2IGB
 
 | | Crystal Structure of PyrR, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus caldolyticus, UMP-bound form | | Descriptor: | 1,2-ETHANEDIOL, PyrR bifunctional protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Chander, P, Switzer, R.L, Smith, J.L. | | Deposit date: | 2006-09-22 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PyrR, the regulator of the pyrimidine biosynthetic operon in Bacillus caldolyticus
To be Published
|
|
1UN1
 
 | | Xyloglucan endotransglycosylase native structure. | | Descriptor: | GOLD ION, XYLOGLUCAN ENDOTRANSGLYCOSYLASE, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A, Henriksson, H, Denman, S, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1UMZ
 
 | | Xyloglucan endotransglycosylase in complex with the xyloglucan nonasaccharide XLLG. | | Descriptor: | XYLOGLUCAN ENDOTRANSGLYCOSYLASE, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[beta-D-galactopyranose-(1-2)-alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Johansson, P, Brumer, H, Kallas, A.M, Henriksson, H, Denman, S.E, Teeri, T.T, Jones, T.A. | | Deposit date: | 2003-09-03 | | Release date: | 2004-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a Poplar Xyloglucan Endotransglycosylase Reveal Details of Transglycosylation Acceptor Binding
Plant Cell, 16, 2004
|
|
1XZN
 
 | | PYRR, THE REGULATOR OF THE PYRIMIDINE BIOSYNTHETIC OPERON IN BACILLUS CALDOLYTICUS, sulfate-bound form | | Descriptor: | MAGNESIUM ION, PyrR bifunctional protein, SULFATE ION | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
1XZ8
 
 | | Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus caldolyticus, Nucleotide-bound form | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
