2OFO
 
 | | MSrecA-native | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OEP
 
 | | MSrecA-ADP-complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2006-12-31 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2OES
 
 | | MSrecA-native-SSB | | Descriptor: | PHOSPHATE ION, Protein recA | | Authors: | Krishna, R, Rajan Prabu, J, Manjunath, G.P, Datta, S, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2007-01-01 | | Release date: | 2007-06-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA
J.Mol.Biol., 367, 2007
|
|
2NXQ
 
 | | Crystal structure of calcium binding protein 1 from Entamoeba histolytica: a novel arrangement of EF hand motifs | | Descriptor: | ACETATE ION, CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Padhan, N, Alam, N, Gourinath, S. | | Deposit date: | 2006-11-18 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of calcium binding protein-1 from Entamoeba histolytica: A novel arrangement of EF hand motifs.
Proteins, 68, 2007
|
|
1G18
 
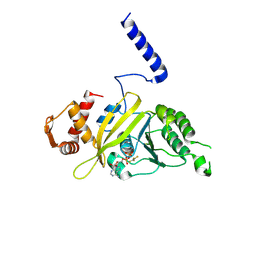 | | RECA-ADP-ALF4 COMPLEX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RECA PROTEIN, TETRAFLUOROALUMINATE ION | | Authors: | Datta, S, Prabu, M.M, Vaze, M.B, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-10-11 | | Release date: | 2000-12-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis RecA and its complex with ADP-AlF(4): implications for decreased ATPase activity and molecular aggregation
Nucleic Acids Res., 28, 2000
|
|
2NWB
 
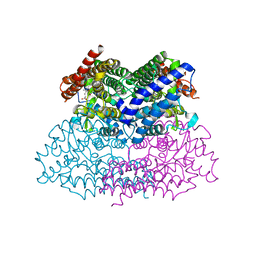 | | Crystal Structure of a Putative 2,3-dioxygenase (SO4414) from Shewanella oneidensis in complex with ferric heme. Northeast Structural Genomics Target SoR52. | | Descriptor: | Conserved domain protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Forouhar, F, Anderson, J.L.R, Mowat, C.G, Hussain, A, Seetharaman, J, Bruckmann, C, Thackray, S.J, Khan, N, Cunningham, K, Janjua, H, Zhao, L, Xiao, R, Ma, L.C, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Champman, S.K, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1M6I
 
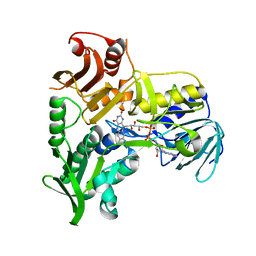 | | Crystal Structure of Apoptosis Inducing Factor (AIF) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Programmed cell death protein 8 | | Authors: | Ye, H, Cande, C, Stephanou, N.C, Jiang, S, Gurbuxani, S, Larochette, N, Daugas, E, Garrido, C, Kroemer, G, Wu, H. | | Deposit date: | 2002-07-16 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DNA binding is required for the apoptogenic action of apoptosis inducing factor.
Nat.Struct.Biol., 9, 2002
|
|
4LM5
 
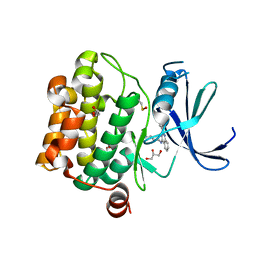 | | Crystal structure of Pim1 in complex with 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol (resulting from displacement of SKF86002) | | Descriptor: | 2-{4-[(3-aminopropyl)amino]quinazolin-2-yl}phenol, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-10 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUD
 
 | | Crystal Structure of HCK in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LUE
 
 | | Crystal Structure of HCK in complex with 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (resulting from displacement of SKF86002) | | Descriptor: | 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LMU
 
 | | Crystal structure of Pim1 in complex with the inhibitor Quercetin (resulting from displacement of SKF86002) | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | Descriptor: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | Authors: | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
