6LNM
 
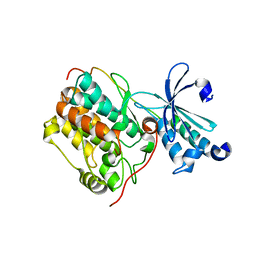 | | Crystal structure of CASK-CaMK in complex with Mint1-CID | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, Peripheral plasma membrane protein CASK | | Authors: | Cai, Q, Zhang, M. | | Deposit date: | 2019-12-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the High-Affinity Interaction between CASK and Mint1.
Structure, 28, 2020
|
|
1IOU
 
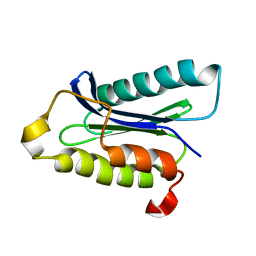 | |
1C1H
 
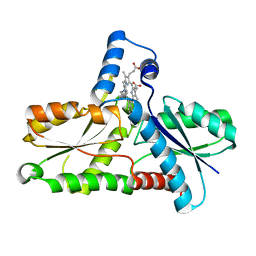 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS FERROCHELATASE IN COMPLEX WITH N-METHYL MESOPORPHYRIN | | Descriptor: | FERROCHELATASE, MAGNESIUM ION, N-METHYLMESOPORPHYRIN | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
6JK0
 
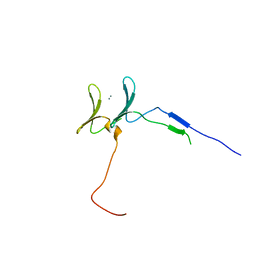 | | Crystal Structure of YAP1 and Dendrin complex | | Descriptor: | CALCIUM ION, Transcriptional coactivator YAP1,Dendrin | | Authors: | Lin, Z, Yang, Z, Ji, Z, Zhang, M. | | Deposit date: | 2019-02-27 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity.
Elife, 8, 2019
|
|
7YUY
 
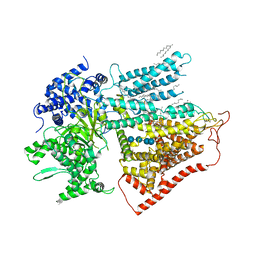 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
3GOQ
 
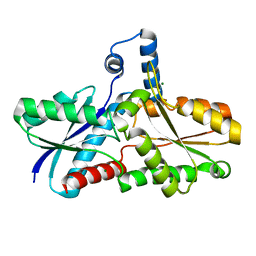 | |
3H91
 
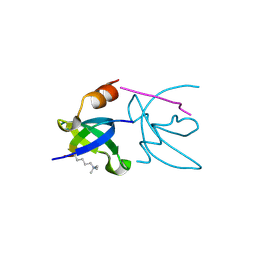 | | Crystal structure of the complex of human chromobox homolog 2 (CBX2) and H3K27 peptide | | Descriptor: | Chromobox protein homolog 2, H3K27 peptide | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2ROW
 
 | | The C1 domain of ROCK II | | Descriptor: | Rho-associated protein kinase 2, ZINC ION | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The C1 domain of ROCK II
To be Published
|
|
3M4Z
 
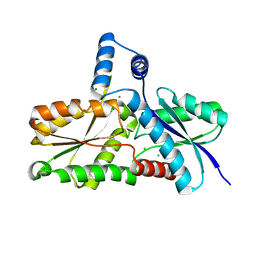 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
6MFW
 
 | | Crystal structure of a 4-domain construct of LgrA in the substrate donation state | | Descriptor: | (2~{R})-~{N}-[3-[2-[[(2~{S})-2-formamido-3-methyl-butanoyl]amino]ethylamino]-3-oxidanylidene-propyl]-3,3-dimethyl-2-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-butanamide, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, Linear gramicidin synthase subunit A, ... | | Authors: | Reimer, J.M, Eivaskhani, M, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
1DOZ
 
 | | CRYSTAL STRUCTURE OF FERROCHELATASE | | Descriptor: | FERROCHELATASE, MAGNESIUM ION | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-12-22 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
2RLO
 
 | | Split PH domain of PI3-kinase enhancer | | Descriptor: | Centaurin-gamma 1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2007-07-21 | | Release date: | 2008-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Split pleckstrin homology domain-mediated cytoplasmic-nuclear localization of PI3-kinase enhancer GTPase
J.Mol.Biol., 378, 2008
|
|
2PKU
 
 | | Solution structure of PICK1 PDZ in complex with the carboxyl tail peptide of GluR2 | | Descriptor: | PRKCA-binding protein, peptide (GLU)(SER)(VAL)(LYS)(ILE) | | Authors: | Pan, L, Wu, H, Shen, C, Shi, Y, Xia, J, Zhang, M. | | Deposit date: | 2007-04-18 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes
Embo J., 26, 2007
|
|
4HGD
 
 | | Structural insights into yeast Nit2: C169S mutant of yeast Nit2 in complex with an endogenous peptide-like ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, N-(4-carboxy-4-oxobutanoyl)-L-cysteinylglycine, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-08 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6KG7
 
 | | Cryo-EM Structure of the Mammalian Tactile Channel Piezo2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Piezo-type mechanosensitive ion channel component 2 | | Authors: | Wang, L, Zhou, H, Zhang, M, Liu, W, Deng, T, Zhao, Q, Li, Y, Lei, J, Li, X, Xiao, B. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and mechanogating of the mammalian tactile channel PIEZO2.
Nature, 573, 2019
|
|
2WCO
 
 | | Structures of the Streptomyces coelicolor A3(2) Hyaluronan Lyase in Complex with Oligosaccharide Substrates and an Inhibitor | | Descriptor: | 4-deoxy-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, GLYCEROL, ... | | Authors: | Elmabrouk, Z.H, Taylor, E.J, Vincent, F, Smith, N.L, Zhang, M, Charnock, S.J, Turkenburg, J.P, Davies, G.J, Black, G.W. | | Deposit date: | 2009-03-12 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of a Family 8 Polysaccharide Lyase Reveal Open and Highly Occluded Substrate-Binding Cleft Conformations.
Proteins, 79, 2011
|
|
1LBQ
 
 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
4HG3
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 in complex with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CACODYLATE ION, GLYCEROL, ... | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-10-06 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4H5U
 
 | | Structural insights into yeast Nit2: wild-type yeast Nit2 | | Descriptor: | CACODYLATE ION, GLYCEROL, Probable hydrolase NIT2 | | Authors: | Liu, H, Qiu, X, Zhang, M, Gao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-09-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of enzyme-intermediate complexes of yeast Nit2: insights into its catalytic mechanism and different substrate specificity compared with mammalian Nit2
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6MFY
 
 | | Crystal structure of a 5-domain construct of LgrA in the substrate donation state | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Linear gramicidin synthase subunit A, PHOSPHATE ION | | Authors: | Reimer, J.M, Eivaskhani, M, Harb, I, Schmeing, T.M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility.
Science, 366, 2019
|
|
6M3P
 
 | | Crystal structure of AnkG/beta2-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain, non-erythrocytic 1 | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
1D1P
 
 | | CRYSTAL STRUCTURE OF A YEAST LOW MOLECULAR WEIGHT PROTEIN TYROSINE PHOSPHATASE (LTP1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, TYROSINE PHOSPHATASE | | Authors: | Wang, S, Tabernero, L, Zhang, M, Harms, E, Van Etten, R.L, Stauffacher, C.V. | | Deposit date: | 1999-09-20 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of a low-molecular weight protein tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate.
Biochemistry, 39, 2000
|
|
2PON
 
 | | Solution structure of the Bcl-xL/Beclin-1 complex | | Descriptor: | Apoptosis regulator Bcl-X, Beclin-1 | | Authors: | Feng, W, Huang, S, Wu, H, Zhang, M. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Bcl-xL's Target Recognition Versatility Revealed by the Structure of Bcl-xL in Complex with the BH3 Domain of Beclin-1.
J.Mol.Biol., 372, 2007
|
|
2QT5
 
 | | Crystal Structure of GRIP1 PDZ12 in Complex with the Fras1 Peptide | | Descriptor: | (ASN)(ASN)(LEU)(GLN)(ASP)(GLY)(THR)(GLU)(VAL), 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Long, J, Wei, Z, Feng, W, Zhao, Y, Zhang, M. | | Deposit date: | 2007-08-01 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Supramodular nature of GRIP1 revealed by the structure of its PDZ12 tandem in complex with the carboxyl tail of Fras1.
J.Mol.Biol., 375, 2008
|
|
4FYP
 
 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
