4FZO
 
 | |
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
2HAP
 
 | | STRUCTURE OF A HAP1-18/DNA COMPLEX REVEALS THAT PROTEIN/DNA INTERACTIONS CAN HAVE DIRECT ALLOSTERIC EFFECTS ON TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), PROTEIN (HEME ACTIVATOR PROTEIN), ... | | Authors: | King, D.A, Zhang, L, Guarente, L, Marmorstein, R. | | Deposit date: | 1998-09-17 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation.
Nat.Struct.Biol., 6, 1999
|
|
5YVF
 
 | | Crystal structure of BFA1 | | Descriptor: | BFA1 | | Authors: | Pu, H, Zhang, L, Duan, Z.K, Peng, L.W, Liu, L. | | Deposit date: | 2017-11-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor 1.
Plant Cell, 30, 2018
|
|
5ZAT
 
 | | Crystal structure of 5-carboxylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(CAC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
5Z6P
 
 | | The crystal structure of an agarase, AgWH50C | | Descriptor: | B-agarase | | Authors: | Mao, X, Zhou, J, Zhang, P, Zhang, L, Zhang, J, Li, Y. | | Deposit date: | 2018-01-24 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Structure-based design of agarase AgWH50C from Agarivorans gilvus WH0801 to enhance thermostability.
Appl. Microbiol. Biotechnol., 103, 2019
|
|
8W72
 
 | |
8W6Z
 
 | | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins | | Descriptor: | (E)-3-[2-[(2R,3S)-3-[(1R)-1-aminocarbonyloxypropyl]oxiran-2-yl]phenyl]prop-2-enoic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fang, C, Zhang, L, Zhu, Y, Zhang, C. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-bound crystal structure of a P450 enzyme DmlH that catalyze intramolecular phenol coupling in the biosynthesis of cihanmycins
To Be Published
|
|
8IF8
 
 | | Arabinosyltransferase AftA | | Descriptor: | CALCIUM ION, Galactan 5-O-arabinofuranosyltransferase | | Authors: | Gong, Y.C, Rao, Z.H, Zhang, L. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the priming arabinosyltransferase AftA required for AG biosynthesis of Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5YY5
 
 | | Structural definition of a unique neutralization epitope on the receptor-binding domain of MERS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, Light chain, ... | | Authors: | Zhang, S, Wang, P, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2017-12-08 | | Release date: | 2018-08-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Definition of a Unique Neutralization Epitope on the Receptor-Binding Domain of MERS-CoV Spike Glycoprotein
Cell Rep, 24, 2018
|
|
2BKL
 
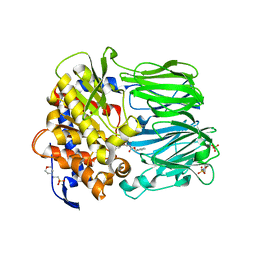 | | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Inter-Domain Dynamics in Catalysis and Specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, N-[(BENZYLOXY)CARBONYL]-L-ALANYL-L-PROLINE, PROLYL ENDOPEPTIDASE, ... | | Authors: | Khosla, C, Shan, L, Mathews, I.I. | | Deposit date: | 2005-02-16 | | Release date: | 2005-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Mechanistic Analysis of Two Prolyl Endopeptidases: Role of Interdomain Dynamics in Catalysis and Specificity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5ZAS
 
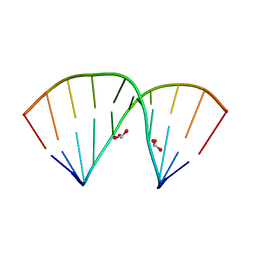 | | Crystal structure of 5-formylcytosine containing decamer dsDNA | | Descriptor: | BICARBONATE ION, DNA (5'-D(*CP*CP*AP*GP*(5FC)P*GP*CP*TP*GP*G)-3') | | Authors: | Fu, T.R, Zhang, L. | | Deposit date: | 2018-02-08 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
4FZP
 
 | |
5IDK
 
 | |
1L0L
 
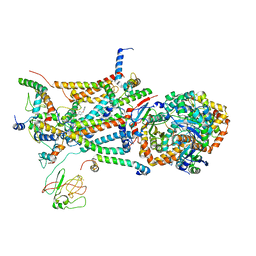 | | structure of bovine mitochondrial cytochrome bc1 complex with a bound fungicide famoxadone | | Descriptor: | Cytochrome B, Cytochrome c1, heme protein, ... | | Authors: | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | Deposit date: | 2002-02-11 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
1L0N
 
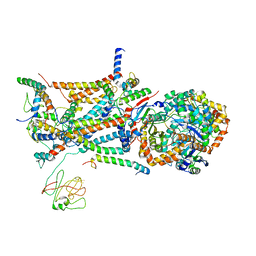 | | native structure of bovine mitochondrial cytochrome bc1 complex | | Descriptor: | Cytochrome B, Cytochrome c1, heme protein, ... | | Authors: | Gao, X, Wen, X, Yu, C.A, Esser, L, Tsao, S, Quinn, B, Zhang, L, Yu, L, Xia, D. | | Deposit date: | 2002-02-11 | | Release date: | 2003-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Mitochondrial Cytochrome bc1 in Complex with Famoxadone: The Role of Aromatic-Aromatic Interaction in Inhibition
Biochemistry, 41, 2003
|
|
2D3Z
 
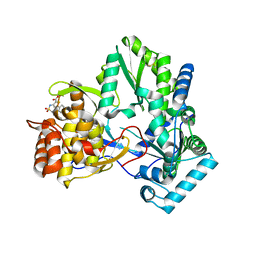 | | X-ray crystal structure of hepatitis C virus RNA-dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-FLUOROPHENYL)-3-{[(4-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-08-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
2D3U
 
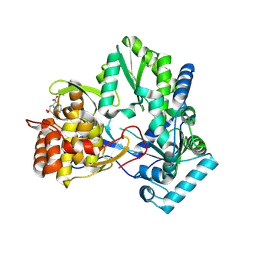 | | X-ray crystal structure of hepatitis C virus RNA dependent RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | 5-(4-CYANOPHENYL)-3-{[(2-METHYLPHENYL)SULFONYL]AMINO}THIOPHENE-2-CARBOXYLIC ACID, polyprotein | | Authors: | Biswal, B.K, Wang, M, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Bedard, J, James, M.N.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-08-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-nucleoside Inhibitors Binding to Hepatitis C Virus NS5B Polymerase Reveal a Novel Mechanism of Inhibition
J.Mol.Biol., 361, 2006
|
|
5MWV
 
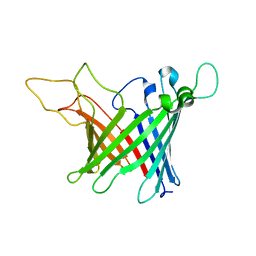 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
5MTK
 
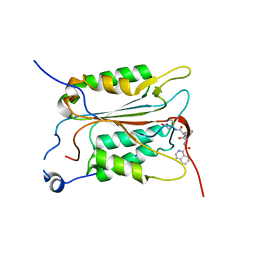 | | Crystal structure of human Caspase-1 with (3S,6S,10aS)-N-((2S,3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-6-(isoquinoline-1-carboxamido)-5-oxodecahydropyrrolo[1,2-a]azocine-3-carboxamide (PGE-3935199) | | Descriptor: | (3~{S})-3-[[(3~{S},6~{S},10~{a}~{S})-6-(isoquinolin-1-ylcarbonylamino)-5-oxidanylidene-2,3,6,7,8,9,10,10~{a}-octahydro-1~{H}-pyrrolo[1,2-a]azocin-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2017-01-09 | | Release date: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Playing against the odds: scaffold hopping from 3D-fragments
To Be Published
|
|
8EYZ
 
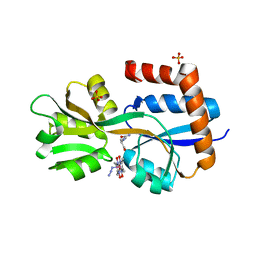 | | Engineered glutamine binding protein bound to GLN and a cobaloxime ligand | | Descriptor: | AZIDOBIS (DIMETHYLGLYOXIMATO) PYRIDINECOBALT, Amino acid ABC transporter substrate-binding protein, GLUTAMINE, ... | | Authors: | Bridwell-Rabb, J, Boggs, D.G, Olshansky, L, Fatima, S, Thompson, P. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Engineering a Conformationally Switchable Artificial Metalloprotein.
J.Am.Chem.Soc., 144, 2022
|
|
5MMV
 
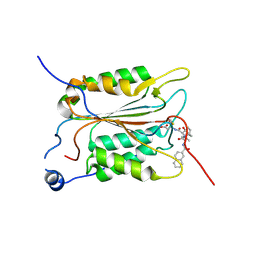 | | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1) | | Descriptor: | (3~{S})-3-[[(3~{S})-2-[(2~{S})-3-methyl-2-(naphthalen-2-ylcarbonylamino)butanoyl]-4-oxidanyl-2-azabicyclo[2.2.2]octan-3-yl]carbonylamino]-4-oxidanyl-butanoic acid, Caspase-1 | | Authors: | Brethon, A, Chantalat, L, Christin, O, Clary, L, Fournier, J.F, Gastreich, M, Harris, C, Pascau, J, Isabet, T, Rodeschin, V, Thoreau, E, Roche, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human Caspase-1 with 2-((2-naphthoyl)-L-valyl)-4-hydroxy-N-((3S)-2-hydroxy-5-oxotetrahydrofuran-3-yl)-2-azabicyclo[2.2.2]octane-3-carboxamide (Compound 1)
To Be Published
|
|
5Z9X
 
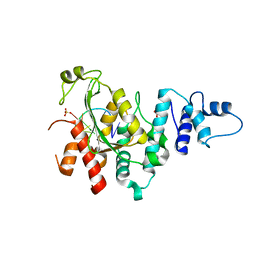 | | Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 in complex with an RNA substrate | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*GP*CP*CP*CP*AP*UP*UP*AP*G)-3'), SULFATE ION, ... | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Gan, J, Cao, C, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
5YVD
 
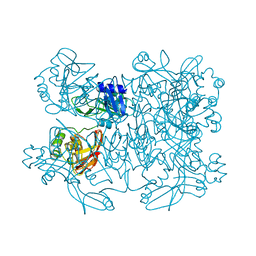 | |
5Z9Z
 
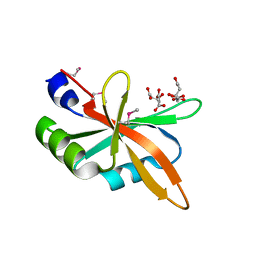 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
