3OQT
 
 | | Crystal structure of Rv1498A protein from mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, Rv1498A PROTEIN, SODIUM ION | | Authors: | Liu, F, Xiong, J, Kumar, S, Yang, C, Li, S, Ge, S, Xia, N, Swaminathan, K. | | Deposit date: | 2010-09-04 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A.
J.Struct.Biol., 175, 2011
|
|
7JVX
 
 | | Crystal structure of PTEN (aa 7-353 followed by spacer TGGGSGGTGGGSGGTGGGCY ligated to peptide pSDpTpTDpSDPENEPFDED) | | Descriptor: | PHOSPHATE ION, Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-24 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3EWE
 
 | | Crystal Structure of the Nup85/Seh1 Complex | | Descriptor: | Nucleoporin NUP85, Nucleoporin SEH1 | | Authors: | Brohawn, S.G, Leksa, N.C, Rajashankar, K.R, Schwartz, T.U. | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Science, 322, 2008
|
|
7JUL
 
 | | Crystal structure of non phosphorylated PTEN (n-crPTEN-13sp-T1, SDTTDSDPENEG) | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN | | Authors: | Dempsey, D, Phan, K, Cole, P, Gabelli, S.B. | | Deposit date: | 2020-08-20 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structural basis of PTEN regulation by multi-site phosphorylation.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2G28
 
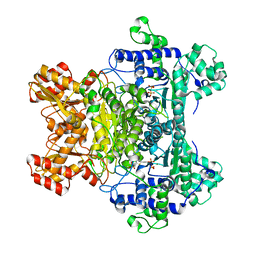 | | E. Coli Pyruvate Dehydrogenase H407A variant Phosphonolactylthiamin Diphosphate Complex | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Furey, W, Arjunan, P, Chandrasekhar, K. | | Deposit date: | 2006-02-15 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Thiamin-bound, Pre-decarboxylation Reaction Intermediate Analogue in the Pyruvate Dehydrogenase E1 Subunit Induces Large Scale Disorder-to-Order Transformations in the Enzyme and Reveals Novel Structural Features in the Covalently Bound Adduct.
J.Biol.Chem., 281, 2006
|
|
3QE5
 
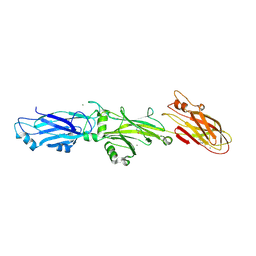 | | Complete structure of Streptococcus mutans Antigen I/II carboxy-terminus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Major cell-surface adhesin PAc, ... | | Authors: | Larson, M.R, Rajashankar, K.R, Crowley, P.J, Kelly, C, Mitchell, T.J, Brady, L.J, Deivanayagam, C. | | Deposit date: | 2011-01-19 | | Release date: | 2011-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the C-terminal Region of Streptococcus mutans Antigen I/II and Characterization of Salivary Agglutinin Adherence Domains.
J.Biol.Chem., 286, 2011
|
|
3FEF
 
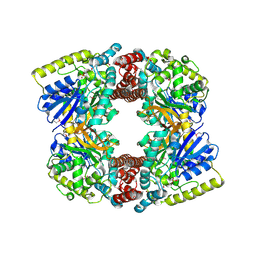 | | Crystal structure of putative glucosidase lplD from bacillus subtilis | | Descriptor: | MAGNESIUM ION, Putative glucosidase lplD, ALPHA-GALACTURONIDASE, ... | | Authors: | Ramagopal, U.A, Rajashankar, K.R, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-28 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glucosidase lplD from bacillus subtilis.
To be published
|
|
3HAJ
 
 | | Crystal structure of human PACSIN2 F-BAR domain (p212121 lattice) | | Descriptor: | CALCIUM ION, human PACSIN2 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2G67
 
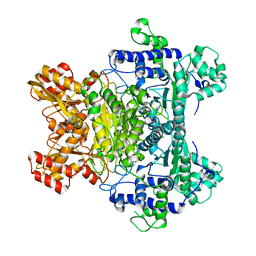 | |
1RP7
 
 | | E. COLI PYRUVATE DEHYDROGENASE INHIBITOR COMPLEX | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Arjunan, P, Chandrasekhar, K, Furey, W. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Determinants of Enzyme Binding Affinity: The E1 Component of Pyruvate Dehydrogenase from Escherichia coli in Complex with the Inhibitor Thiamin Thiazolone Diphosphate.
Biochemistry, 43, 2004
|
|
3HAH
 
 | | Crystal structure of human PACSIN1 F-BAR domain (C2 lattice) | | Descriptor: | CALCIUM ION, human PACSIN1 F-BAR | | Authors: | Wang, Q, Navarro, M.V.A.S, Peng, G, Rajashankar, K.R, Sondermann, H. | | Deposit date: | 2009-05-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6A4S
 
 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | Descriptor: | Peptidase E | | Authors: | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
2Z9Y
 
 | | Crystal structure of CERT START domain in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3FOD
 
 | |
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
1THS
 
 | | STRUCTURES OF THROMBIN COMPLEXES WITH A DESIGNED AND A NATURAL EXOSITE INHIBITOR | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), SYNTHETIC INHIBITOR | | Authors: | Qiu, X, Yin, M, Padmanabhan, K.P, Krstenansky, J.L, Tulinsky, A. | | Deposit date: | 1993-06-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of thrombin complexes with a designed and a natural exosite peptide inhibitor.
J.Biol.Chem., 268, 1993
|
|
1BKU
 
 | | EFFECTS OF GLYCOSYLATION ON THE STRUCTURE AND DYNAMICS OF EEL CALCITONIN, NMR, 10 STRUCTURES | | Descriptor: | CALCITONIN | | Authors: | Hashimoto, Y, Nishikido, J, Toma, K, Yamamoto, K, Haneda, K, Inazu, T, Valentine, K, Opella, S.J. | | Deposit date: | 1998-07-13 | | Release date: | 1999-01-13 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Effects of glycosylation on the structure and dynamics of eel calcitonin in micelles and lipid bilayers determined by nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
3JS2
 
 | | Crystal structure of minimal kinase domain of fibroblast growth factor receptor 1 in complex with 5-(2-thienyl)nicotinic acid | | Descriptor: | 5-(2-thienyl)nicotinic acid, Basic fibroblast growth factor receptor 1, PHOSPHATE ION | | Authors: | Bae, J.H, Ravindranathan, K.P, Mandiyan, V, Ekkati, A.R, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
J.Med.Chem., 53, 2010
|
|
3BC9
 
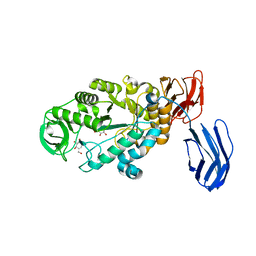 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
1U79
 
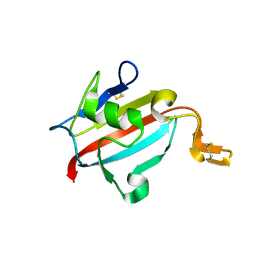 | | Crystal structure of AtFKBP13 | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Gopalan, G, Swaminathan, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
3BCD
 
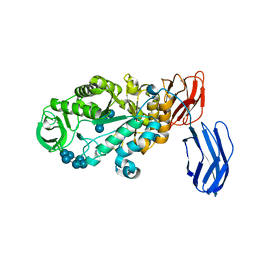 | | Alpha-amylase B in complex with maltotetraose and alpha-cyclodextrin | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCF
 
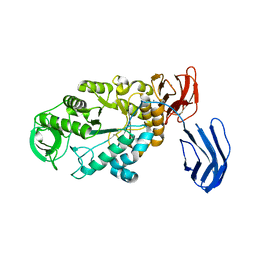 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
4NX5
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-AZA URIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-12-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate
To be Published
|
|
4IBL
 
 | | Rubidium Sites in Blood Coagulation Factor VIIa | | Descriptor: | BENZAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vadivel, K, Schmidt, A, Cascio, D, Padmanabhan, K, Bajaj, S.P. | | Deposit date: | 2012-12-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human factor VIIa-soluble tissue factor with calcium, magnesium and rubidium
Acta Crystallogr.,Sect.D, D77, 2021
|
|
