4ZLK
 
 | | Crystal structure of mouse myosin-5a in complex with calcium-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Unconventional myosin-Va | | Authors: | Shen, M, Zhang, N, Zheng, S, Zhang, W.-B, Zhang, H.-M, Lu, Z, Su, Q.P, Sun, Y, Ye, K, Li, X.-D. | | Deposit date: | 2015-05-01 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structural basis for calcium regulation of myosin 5 motor function
To Be Published
|
|
4K3C
 
 | | The crystal structure of BamA from Haemophilus ducreyi lacking POTRA domains 1-3 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
4K3B
 
 | | The crystal structure of BamA from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
5V7R
 
 | |
5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
6SPF
 
 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPE
 
 | | Pseudomonas aeruginosa 30s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPC
 
 | | Pseudomonas aeruginosa 30s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4XWO
 
 | | Structure of Get3 bound to the transmembrane domain of Sec22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATPase GET3, ... | | Authors: | Mateja, A, Paduch, M, Chang, H.-Y, Szydlowska, A, Kossiakoff, A.A, Hegde, R.S, Keenan, R.J. | | Deposit date: | 2015-01-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo.
Science, 347, 2015
|
|
5VJX
 
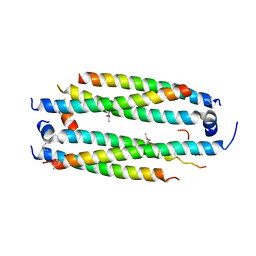 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
5VJI
 
 | | Crystal structure of the CLOCK Transcription Domain Exon19 in Complex with a Repressor | | Descriptor: | CLOCK-interacting pacemaker, Circadian locomoter output cycles protein kaput | | Authors: | Hou, Z, Su, L, Pei, J, Grishin, N.V, Zhang, H. | | Deposit date: | 2017-04-19 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the CLOCK Transactivation Domain Exon19 in Complex with a Repressor.
Structure, 25, 2017
|
|
6WAP
 
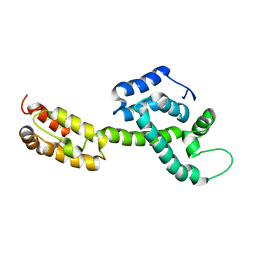 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5V7U
 
 | |
6SPB
 
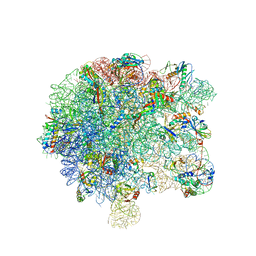 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate with a mutation in uL6 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPD
 
 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6SPG
 
 | | Pseudomonas aeruginosa 70s ribosome from a clinical isolate | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7EXA
 
 | | Structure of mumps virus nucleoprotein without C-arm | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N, Qin, Y. | | Deposit date: | 2021-05-26 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural plasticity of mumps virus nucleocapsids with cryo-EM structures.
Commun Biol, 4, 2021
|
|
5E6N
 
 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5H15
 
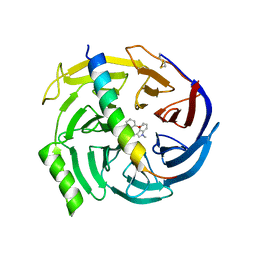 | | EED in complex with PRC2 allosteric inhibitor EED709 | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5WSZ
 
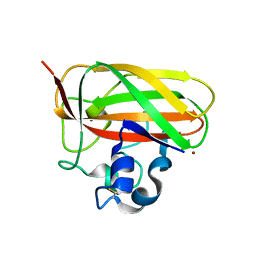 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
7ESF
 
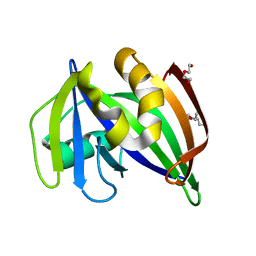 | | The Crystal Structure of human MTH1 from Biortus | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Wang, F, Cheng, W, Shang, H, Wang, R, Zhang, B, Tian, F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Crystal Structure of human MTH1 from Biortus
To Be Published
|
|
7XW9
 
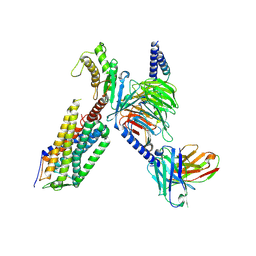 | | Cryo-EM structure of the TRH-bound human TRHR-Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha, ... | | Authors: | Ji, S, Dong, Y, Chen, L, Zang, S, Shen, D, Guo, J, Qin, J, Zhang, H, Wang, W, Shen, Q, Mao, C, Zhang, Y. | | Deposit date: | 2022-05-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis for the activation of thyrotropin-releasing hormone receptor.
Cell Discov, 8, 2022
|
|
5GQQ
 
 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
7XWO
 
 | | Neurokinin A bound to active human neurokinin 2 receptor in complex with G324 | | Descriptor: | G324, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, W.J, Yuan, Q.N, Zhang, H.H, Yang, F, Ling, S.L, Lv, P, Eric, X, Tian, C.L, Yin, W.C, Shi, P. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the activation of neurokinin 2 receptor by neurokinin A.
Cell Discov, 8, 2022
|
|
