8DGC
 
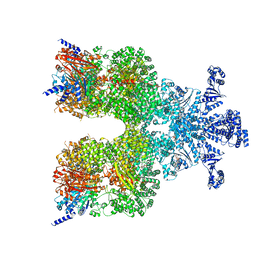 | | Avs3 bound to phage PhiV-1 terminase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SeAvs3, ... | | Authors: | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
8E4L
 
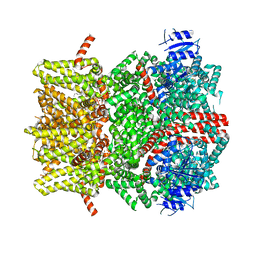 | | The open state mouse TRPM8 structure in complex with the cooling agonist C3, AITC, and PI(4,5)P2 | | Descriptor: | 3-isothiocyanatoprop-1-ene, CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4O
 
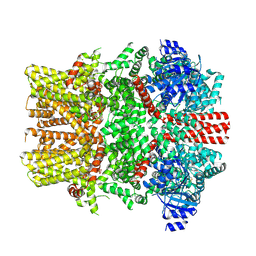 | | The closed C1-state mouse TRPM8 structure in complex with putative PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4N
 
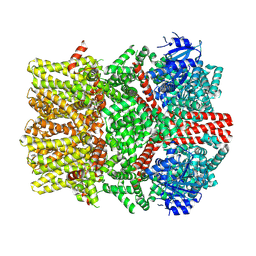 | | The closed C1-state mouse TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4Q
 
 | | The closed C0-state flycatcher TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4P
 
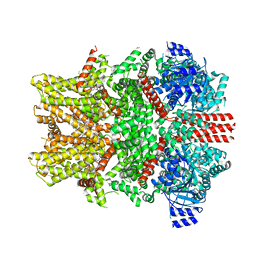 | | Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class I , C0 state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4M
 
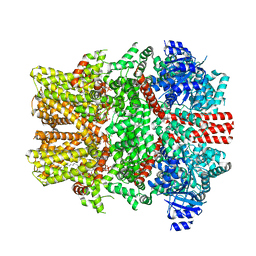 | | The intermediate C2-state mouse TRPM8 structure in complex with the cooling agonist C3 and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
1EIO
 
 | | ILEAL LIPID BINDING PROTEIN IN COMPLEX WITH GLYCOCHOLATE | | Descriptor: | GLYCOCHOLIC ACID, ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Hamilton, J.A, Sacchettini, J.C, Rueterjans, H. | | Deposit date: | 2000-02-27 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ileal lipid binding protein in complex with glycocholate.
Eur.J.Biochem., 267, 2000
|
|
4U63
 
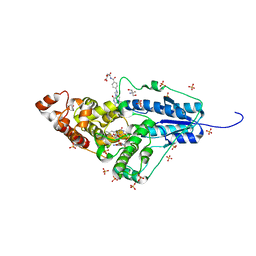 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
1EAL
 
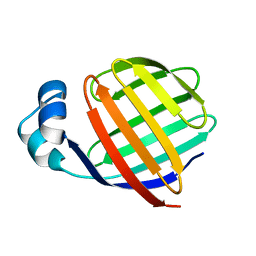 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
6DKL
 
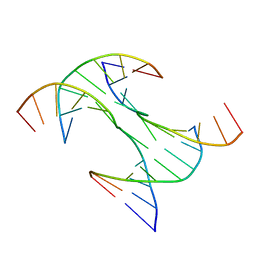 | | Crystal Structure of a Rationally Designed Six-Fold Symmetric DNA Scaffold | | Descriptor: | DNA (5'-D(P*CP*AP*CP*AP*CP*CP*GP*TP*AP*C)-3'), DNA (5'-D(P*GP*GP*AP*TP*GP*CP*AP*CP*A)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*GP*AP*TP*CP*CP*AP*G)-3'), ... | | Authors: | Simmons, C.R, Zhang, F, Yan, H. | | Deposit date: | 2018-05-29 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.034 Å) | | Cite: | Self-Assembly of a 3D DNA Crystal Structure with Rationally Designed Six-Fold Symmetry.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7LPE
 
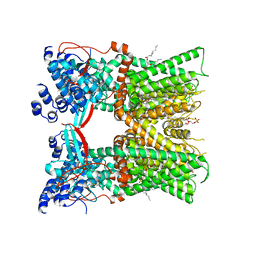 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an open state, class 1 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPA
 
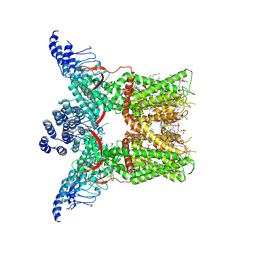 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 4 degrees Celsius | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPB
 
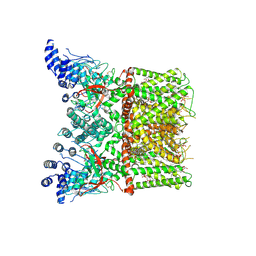 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 25 degrees Celsius | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPD
 
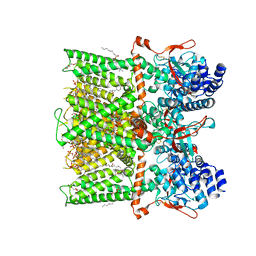 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an intermediate state, class 2 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LP9
 
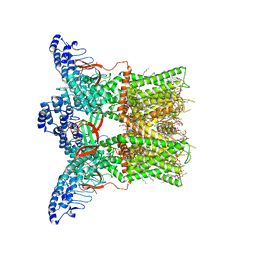 | | Cryo-EM structure of full-length TRPV1 at 4 degrees Celsius | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, Phosphatidylinositol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPC
 
 | | Cryo-EM structure of full-length TRPV1 at 48 degrees Celsius | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, Phosphatidylinositol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3H4M
 
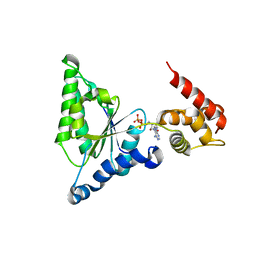 | | AAA ATPase domain of the proteasome- activating nucleotidase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Proteasome-activating nucleotidase | | Authors: | Jeffrey, P, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.106 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
3H43
 
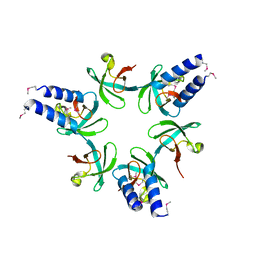 | | N-terminal domain of the proteasome-activating nucleotidase of Methanocaldococcus jannaschii | | Descriptor: | Proteasome-activating nucleotidase | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-17 | | Release date: | 2009-06-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
6UJV
 
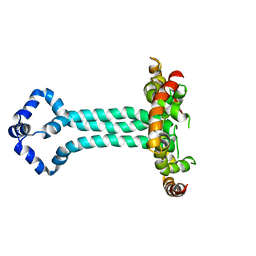 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
5KCM
 
 | | Crystal structure of iron-sulfur cluster containing photolyase PhrB mutant I51W | | Descriptor: | (6-4) photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, X, Bowatte, K, Zhang, F, Lamparter, T. | | Deposit date: | 2016-06-06 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
8FN4
 
 | | Cryo-EM structure of RNase-treated RESC-A in trypanosomal RNA editing | | Descriptor: | RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), RNA-editing substrate-binding complex protein 3 (RESC3), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-26 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FNC
 
 | | Cryo-EM structure of RNase-treated RESC-C in trypanosomal RNA editing | | Descriptor: | Mitochondrial RNA binding complex 1 subunit, Mitochondrial RNA binding protein, Phytanoyl-CoA dioxygenase family protein, ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
8FN6
 
 | | Cryo-EM structure of RNase-untreated RESC-A in trypanosomal RNA editing | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RNA-editing substrate-binding complex protein 1 (RESC1), RNA-editing substrate-binding complex protein 2 (RESC2), ... | | Authors: | Liu, S, Wang, H, Li, X, Zhang, F, Lee, J.K.J, Li, Z, Yu, C, Zhao, X, Hu, J.J, Suematsu, T, Alvarez-Cabrera, A.L, Liu, Q, Zhang, L, Huang, L, Aphasizheva, I, Aphasizhev, R, Zhou, Z.H. | | Deposit date: | 2022-12-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of gRNA stabilization and mRNA recognition in trypanosomal RNA editing.
Science, 381, 2023
|
|
