3RJT
 
 | | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
2J94
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-1H-1,2,4-TRIAZOLE-3-SULFONAMIDE, CALCIUM ION, COAGULATION FACTOR X | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
4MTN
 
 | |
3SHP
 
 | | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | Putative acetyltransferase Sthe_0691, S,R MESO-TARTARIC ACID | | Authors: | Chang, C, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of putative acetyltransferase from Sphaerobacter thermophilus DSM 20745
To be Published
|
|
3OMD
 
 | | Crystal structure of unknown function protein from Leptospirillum rubarum | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Chen, Z, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of unknown function protein from Leptospirillum rubarum
To be Published
|
|
3OKX
 
 | | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris | | Descriptor: | S-ADENOSYLMETHIONINE, YaeB-like protein RPA0152 | | Authors: | Chang, C, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-25 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YaeB-like protein from Rhodopseudomonas palustris
To be Published
|
|
3OIZ
 
 | | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides | | Descriptor: | Antisigma-factor antagonist, STAS | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of antisigma-factor antagonist, STAS domain from Rhodobacter sphaeroides
To be Published
|
|
3OIO
 
 | | Crystal structure of transcriptional regulator (AraC-type DNA-binding domain-containing proteins) from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, SULFATE ION, Transcriptional regulator (AraC-type DNA-binding domain-containing proteins) | | Authors: | Chang, C, Mack, J, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-19 | | Release date: | 2010-09-08 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of transcriptional regulator (AraC-type DNA-binding domain-containing proteins) from Chromobacterium violaceum
To be Published
|
|
3OMB
 
 | | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | Extracellular solute-binding protein, family 1, MAGNESIUM ION | | Authors: | Chang, C, Xu, X, Chin, S, Cui, H, Dong, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
To be Published
|
|
2FLS
 
 | | Crystal structure of Human Glutaredoxin 2 complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin-2 | | Authors: | Johansson, C, Smee, C, Kavanagh, K.L, Debreczeni, J, von Delft, F, Gileadi, O, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-06 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Human Glutaredoxin 2 complexed with glutathione
To be Published
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | Descriptor: | uncharacterized protein SgcJ | | Authors: | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-20 | | Release date: | 2013-12-25 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.719 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
1IHR
 
 | | Crystal structure of the dimeric C-terminal domain of TonB | | Descriptor: | BROMIDE ION, TonB protein | | Authors: | Chang, C, Mooser, A, Pluckthun, A, Wlodawer, A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the dimeric C-terminal domain of TonB reveals a novel fold.
J.Biol.Chem., 276, 2001
|
|
3HDT
 
 | | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, putative kinase | | Authors: | Chang, C, Freeman, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940
To be Published
|
|
2GS3
 
 | | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4) | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Johansson, C, Kavanagh, K.L, Rojkova, A, Gileadi, O, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the selenocysteine to glycine mutant of human glutathione peroxidase 4(GPX4)
To be Published
|
|
3HSI
 
 | |
3ILX
 
 | |
3IX7
 
 | |
2HO5
 
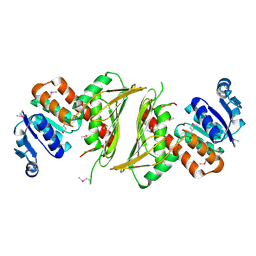 | | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae | | Descriptor: | Oxidoreductase, Gfo/Idh/MocA family | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of Oxidoreductase, Gfo/Idh/MocA family from Streptococcus pneumoniae
To be Published
|
|
3IC6
 
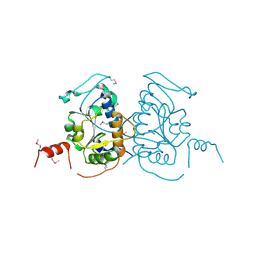 | |
3IH6
 
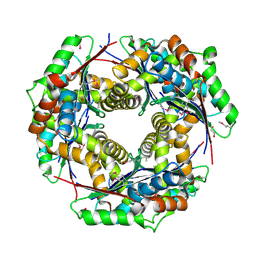 | | Crystal structure of putative zinc protease from Bordetella pertussis Tohama I | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative zinc protease | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative zinc protease from Bordetella pertussis Tohama I
To be Published
|
|
2HT9
 
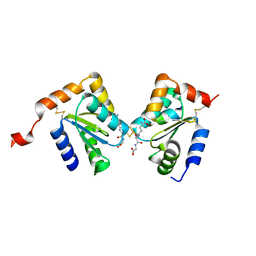 | | The structure of dimeric human glutaredoxin 2 | | Descriptor: | 12-mer peptide, FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, ... | | Authors: | Johansson, C, Smee, C, Kavanagh, K.L, Debreczeni, J, von Delft, F, Gileadi, O, Arrowsmith, C, Weigelt, J, Edwards, A, Sundstrom, M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible sequestration of active site cysteines in a 2Fe-2S-bridged dimer provides a mechanism for glutaredoxin 2 regulation in human mitochondria
J.Biol.Chem., 282, 2007
|
|
3HL0
 
 | | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Maleylacetate reductase, ... | | Authors: | Chang, C, Evdokimova, E, Mursleen, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Maleylacetate reductase from Agrobacterium tumefaciens
To be Published
|
|
1K2A
 
 | | Modified Form of Eosinophil-derived Neurotoxin | | Descriptor: | SULFATE ION, eosinophil-derived neurotoxin | | Authors: | Chang, C, Newton, D.L, Rybak, S.M, Wlodawer, A. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystallographic and functional studies of a modified form of eosinophil-derived neurotoxin (EDN) with novel biological activities.
J.Mol.Biol., 317, 2002
|
|
2I45
 
 | |
3GDW
 
 | | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, Sigma-54 interaction domain protein | | Authors: | Chang, C, Sather, A, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-10 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of sigma-54 interaction domain protein from Enterococcus faecalis
To be Published
|
|
