6XI6
 
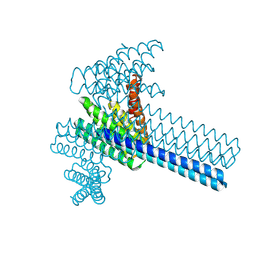 | | Hierarchical design of multi-scale protein complexes by combinatorial assembly of oligomeric helical bundle and repeat protein building blocks | | Descriptor: | helical fusion design | | Authors: | Bera, A.K, Hsia, Y, Kang, A.S, Shankaran, B, Baker, D. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
8HPM
 
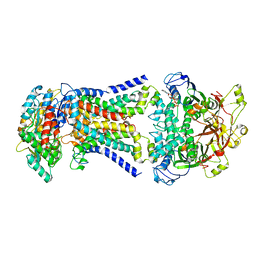 | | LpqY-SugABC in state 2 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPN
 
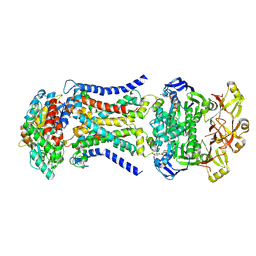 | | LpqY-SugABC in state 3 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPS
 
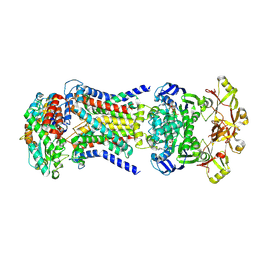 | | LpqY-SugABC in state 5 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
2YOJ
 
 | | HCV NS5B polymerase complexed with pyridonylindole compound | | Descriptor: | 4-fluoranyl-6-[(7-fluoranyl-4-oxidanylidene-3H-quinazolin-6-yl)methyl]-8-(2-oxidanylidene-1H-pyridin-3-yl)furo[2,3-e]indole-7-carboxylic acid, PHOSPHATE ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Chen, K.X, Venkatraman, S, Anilkumar, G.N, Zeng, Q, Lesburg, C.A, Vibulbhan, B, Yang, W, Velazquez, F, Chan, T.-Y, Bennett, F, Sannigrahi, M, Jiang, Y, Duca, J.S, Pinto, P, Gavalas, S, Huang, Y, Wu, W, Selyutin, O, Agrawal, S, Feld, B, Huang, H.-C, Li, C, Cheng, K.-C, Shih, N.-Y, Kozlowski, J.A, Rosenblum, S.B, Njoroge, F.G. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery of Sch 900188: A Potent Hepatitis C Virus Ns5B Polymerase Inhibitor Prodrug as a Development Candidate
Acs Med.Chem.Lett., 5, 2014
|
|
4S1R
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H08.F-117225, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H08.F-117225 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
4S1Q
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
8HPL
 
 | | LpqY-SugABC in state 1 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
8HPR
 
 | | LpqY-SugABC in state 4 | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liang, J, Yang, X, Zhang, B, Rao, Z, Liu, F. | | Deposit date: | 2022-12-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into trehalose capture and translocation by mycobacterial LpqY-SugABC.
Structure, 31, 2023
|
|
4S1S
 
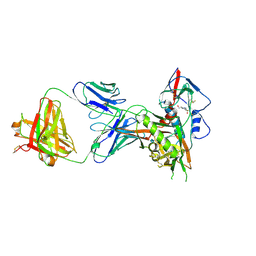 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H5.F-185917, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab of VRC01 light chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
1L3Q
 
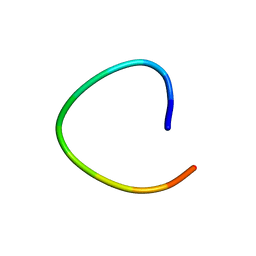 | | H. rufescens abalone shell Lustrin A consensus repeat, FPGKNVNCTSGE, pH 7.4, 1-H NMR structure | | Descriptor: | Lustrin A | | Authors: | Evans, J.S, Wustman, B.A, Zhang, B, Morse, D.E. | | Deposit date: | 2002-02-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Model peptide studies of sequence regions in the elastomeric biomineralization protein, Lustrin A. I. The C-domain consensus-PG-, -NVNCT-motif
Biopolymers, 63, 2002
|
|
8HXQ
 
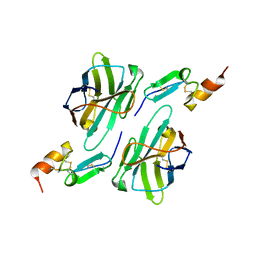 | | Nanobody1 in complex with human BCMA ECD | | Descriptor: | Nanobody1, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
8HXR
 
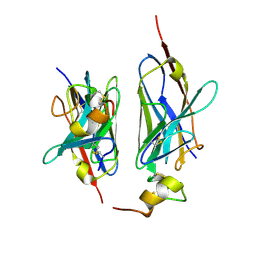 | | Nanobody2 in complex with human BCMA ECD | | Descriptor: | Nanobody2, Tumor necrosis factor receptor superfamily member 17 | | Authors: | Sun, Y, Zhang, B. | | Deposit date: | 2023-01-05 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antigen-induced chimeric antigen receptor multimerization amplifies on-tumor cytotoxicity.
Signal Transduct Target Ther, 8, 2023
|
|
1HX8
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF DROSOPHILA AP180 | | Descriptor: | SULFATE ION, SYNAPSE-ENRICHED CLATHRIN ADAPTOR PROTEIN LAP | | Authors: | Mao, Y, Chen, J, Maynard, J.A, Zhang, B, Quiocho, F.A. | | Deposit date: | 2001-01-12 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel all helix fold of the AP180 amino-terminal domain for phosphoinositide binding and clathrin assembly in synaptic vesicle endocytosis.
Cell(Cambridge,Mass.), 104, 2001
|
|
4GKY
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4201 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
4GKX
 
 | | Crystal structure of a carbohydrate-binding domain | | Descriptor: | CALCIUM ION, GLYCEROL, Protein ERGIC-53, ... | | Authors: | Page, R.C, Zheng, C, Nix, J.C, Misra, S, Zhang, B. | | Deposit date: | 2012-08-13 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Characterization of Carbohydrate Binding by LMAN1 Protein Provides New Insight into the Endoplasmic Reticulum Export of Factors V (FV) and VIII (FVIII).
J.Biol.Chem., 288, 2013
|
|
6A9O
 
 | | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis | | Descriptor: | 2'-[(6-oxo-5,6-dihydrophenanthridin-3-yl)carbamoyl][1,1'-biphenyl]-2-carboxylic acid, DIMETHYL SULFOXIDE, Dihydrogen tetrasulfide, ... | | Authors: | Manjula, R, Padmanabhan, B. | | Deposit date: | 2018-07-14 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational discovery of a SOD1 tryptophan oxidation inhibitor with therapeutic potential for amyotrophic lateral sclerosis.
J.Biomol.Struct.Dyn., 37, 2019
|
|
5JON
 
 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
2KA7
 
 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
5DG4
 
 | | Crystal structure of monomer human cellular retinol binding protein II-Y60L | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Gieger, J.H, Borhan, B. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
4KR0
 
 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
2KL2
 
 | | NMR solution structure of A2LD1 (gi:13879369) | | Descriptor: | AIG2-like domain-containing protein 1 | | Authors: | Pedrini, B, Serrano, P, Mohanty, B, Geralt, M, Herrmann, T, Wuthrich, K, Wilson, I, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Comparison of NMR and crystal structures highlights conformational isomerism in protein active sites.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
7LNA
 
 | | Infectious mammalian prion fibril (263K scrapie) | | Descriptor: | Major prion protein | | Authors: | Kraus, A, Hoyt, F, Schwartz, C.L, Hansen, B, Hughson, A.G, Artikis, E, Race, B, Caughey, B. | | Deposit date: | 2021-02-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution structure and strain comparison of infectious mammalian prions.
Mol.Cell, 81, 2021
|
|
