3K2I
 
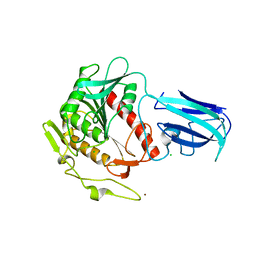 | | Human Acyl-coenzyme A thioesterase 4 | | Descriptor: | Acyl-coenzyme A thioesterase 4, CHLORIDE ION, NICKEL (II) ION | | Authors: | Siponen, M.I, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kraulis, P, Kotenyova, T, Kotzsch, A, Markova, N, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human Acyl-coenzyme A thioesterase 4
To be Published
|
|
4YEH
 
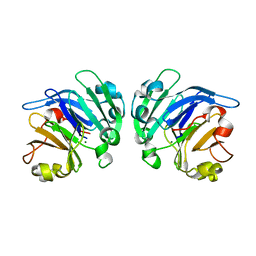 | | Crystal structure of Mg2+ ion containing hemopexin fold from Kabuli chana (chickpea white) at 2.45A resolution reveals a structural basis of metal ion transport | | Descriptor: | Lectin, MAGNESIUM ION | | Authors: | Kumar, S, Singh, A, Yamini, S, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Mg(2+) Containing Hemopexin-Fold Protein from Kabuli Chana (Chickpea-White, CW-25) at 2.45 angstrom Resolution Reveals Its Metal Ion Transport Property
Protein J., 34, 2015
|
|
8GIJ
 
 | |
5NRG
 
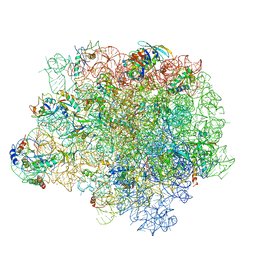 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with RB02 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2017-04-23 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.442 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
6FXC
 
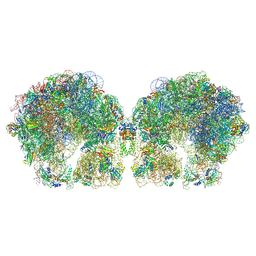 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
3BIN
 
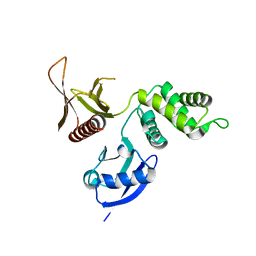 | | Structure of the DAL-1 and TSLC1 (372-383) complex | | Descriptor: | Band 4.1-like protein 3, Cell adhesion molecule 1 | | Authors: | Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Persson, C, Hallberg, B.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B)
J.Biol.Chem., 286, 2011
|
|
3HNG
 
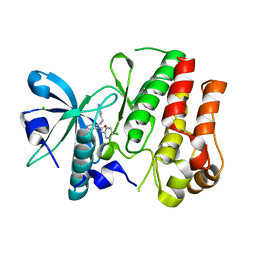 | | Crystal structure of VEGFR1 in complex with N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamide | | Descriptor: | CHLORIDE ION, N-(4-chlorophenyl)-2-[(pyridin-4-ylmethyl)amino]benzamide, Vascular endothelial growth factor receptor 1 | | Authors: | Tresaugues, L, Roos, A, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Kragh-Nielsen, T, Kotzch, A, Sagemark, J, Schueler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van der Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-31 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of VEGFR1 in complex with N-(4-Chlorophenyl)-2-((pyridin-4-ylmethyl)amino)benzamide
To be Published
|
|
4DO1
 
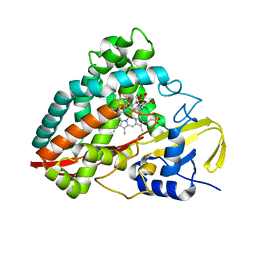 | | The crystal structures of 4-methoxybenzoate bound CYP199A4 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNJ
 
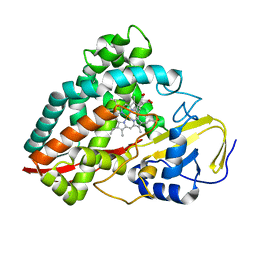 | | The crystal structures of 4-methoxybenzoate bound CYP199A2 | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-08 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
4DNZ
 
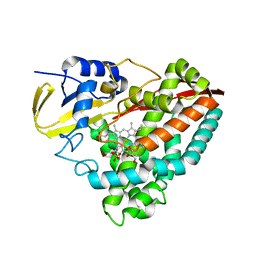 | | The crystal structures of CYP199A4 | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Tan, A.B.H, Zhou, R, Johnson, E.O.D, Zhang, A, Rao, Z, Wong, L.-L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of 4-methoxybenzoate bound CYP199A2 and CYP199A4: structural changes on substrate binding and the identification of an anion binding site
Dalton Trans, 41, 2012
|
|
7Z52
 
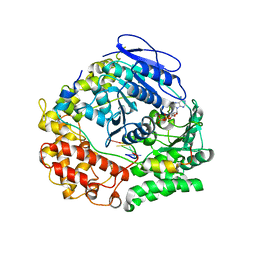 | | Human NEXT dimer - focused reconstruction of the single MTR4 | | Descriptor: | Exosome RNA helicase MTR4, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RNA (5'-R(P*UP*UP*UP*UP*U)-3'), ... | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Z
 
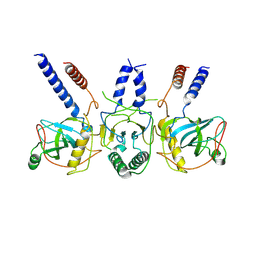 | | Human NEXT dimer - focused reconstruction of the dimerization module | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
7Z4Y
 
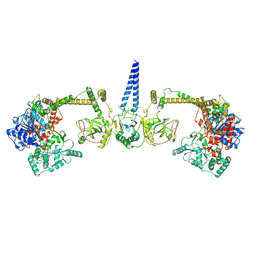 | | Human NEXT dimer - overall reconstruction of the core complex | | Descriptor: | Exosome RNA helicase MTR4, Zinc finger CCHC domain-containing protein 8 | | Authors: | Gerlach, P, Lingaraju, M, Salerno-Kochan, A, Bonneau, F, Basquin, J, Conti, E. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and regulation of the nuclear exosome targeting complex guides RNA substrates to the exosome.
Mol.Cell, 82, 2022
|
|
5HWK
 
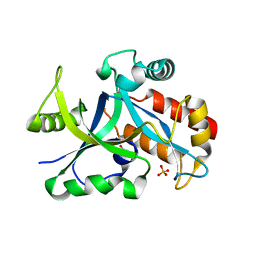 | | Crystal structure of gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | BENZOIC ACID, Glutathione-specific gamma-glutamylcyclotransferase, PHOSPHATE ION | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.344 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
3D8B
 
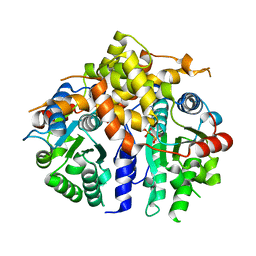 | | Crystal structure of human fidgetin-like protein 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fidgetin-like protein 1 | | Authors: | Karlberg, T, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-23 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human fidgetin-like protein 1.
To be Published
|
|
3M7S
 
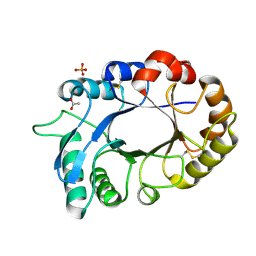 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
5HKV
 
 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lincomycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23s ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Yonath, A, Matzov, D, Eyal, Z, Ben Hamou, R, Zimmerman, E, Rozenberg, H, Bashan, A, Fridman, M. | | Deposit date: | 2016-01-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus.
Nucleic Acids Res., 45, 2017
|
|
3IHJ
 
 | | Human alanine aminotransferase 2 in complex with PLP | | Descriptor: | Alanine aminotransferase 2, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wisniewska, M, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Kotzsch, A, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutamate pyruvate transaminase 2
To be Published
|
|
3E77
 
 | | Human phosphoserine aminotransferase in complex with PLP | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | Authors: | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, S.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Wisniewska, M, Weigelt, J, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human phosphoserine aminotransferase in complex with PLP
TO BE PUBLISHED
|
|
4QD3
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Pseudomonas aeruginosa with 5-azacytidine at 1.89 Angstrom resolution | | Descriptor: | 4-amino-1-(beta-D-ribofuranosyl)-1,3,5-triazin-2(1H)-one, GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Singh, A, Gautam, L, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-13 | | Release date: | 2014-06-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase
Biochem.J., 463, 2014
|
|
3EQ5
 
 | | Crystal structure of fragment 137 to 238 of the human Ski-like protein | | Descriptor: | Ski-like protein | | Authors: | Tresaugues, L, Wisniewska, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Schueler, H, Thorsell, A.G, Van Den Berg, S, Welin, M, Wikstrom, M, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2009-01-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of fragment 137 to 238 of the human Ski-like protein.
To be Published
|
|
4Q7N
 
 | | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, N,N,N-trimethyl-4-oxobutan-1-aminium | | Authors: | Chaudhary, A, Tyagi, T.K, Singh, A, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex of Buffalo Signalling protein SPB-40 with 4-N-trimethylaminobutyraldehyde at 1.79 Angstrom Resolution
To be Published
|
|
5HWI
 
 | | Crystal structure of selenomethionine labelled gama glutamyl cyclotransferease specific to glutathione from yeast | | Descriptor: | GLYCEROL, Glutathione-specific gamma-glutamylcyclotransferase, SUCCINIC ACID | | Authors: | Kaur, A, Gautam, R, Srivastava, R, Chandel, A, Kumar, A, Karthikeyan, S, Bachhawat, A.K. | | Deposit date: | 2016-01-29 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione
J. Biol. Chem., 292, 2017
|
|
6TPH
 
 | |
6AHE
 
 | | Crystal structure of enoyl-ACP reductase from Acinetobacter baumannii in complex with NAD and AFN-1252 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Rani, S.T, Nataraj, V, Laxminarasimhan, A, Thomas, A, Krishnamurthy, N. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Ternary complex formation of AFN-1252 with Acinetobacter baumannii FabI and NADH: Crystallographic and biochemical studies.
Chem.Biol.Drug Des., 96, 2020
|
|
