5K18
 
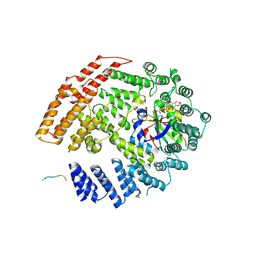 | | The NatB Acetyltransferase Complex Bound To bisubstrate inhibitor | | Descriptor: | Bisubstrate inhibitor, COENZYME A, N-terminal acetyltransferase B complex subunit NAT3, ... | | Authors: | Hong, H, Cai, Y, Zhang, S, Han, A. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular Basis of Substrate Specific Acetylation by N-Terminal Acetyltransferase NatB
Structure, 25, 2017
|
|
4OQO
 
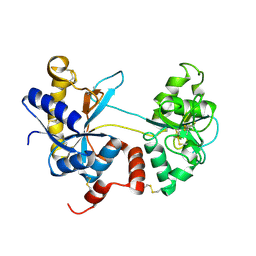 | | Crystal structure of the tryptic generated iron-free C-lobe of bovine Lactoferrin at 2.42 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lactotransferrin | | Authors: | Singh, A, Rastogi, N, Pandey, S, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of the iron-free true C-terminal half of bovine lactoferrin produced by tryptic digestion and its functional significance in the gut.
Febs J., 281, 2014
|
|
3UMQ
 
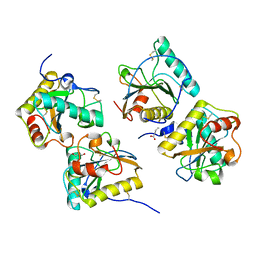 | | Crystal structure of peptidoglycan recognition protein-S complexed with butyric acid at 2.2 A resolution | | Descriptor: | GLYCEROL, Peptidoglycan recognition protein 1, butanoic acid | | Authors: | Pandey, N, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-11-14 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site
Arch.Biochem.Biophys., 529, 2013
|
|
3CXA
 
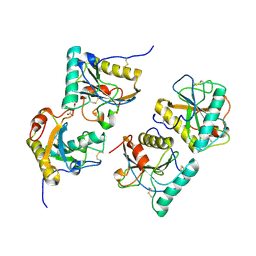 | | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Balaji, K, Sharma, P, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein with alpha-D-glucopyranosyl alpha-D-glucopyranoside at 3.4 A resolution
To be Published
|
|
4OPP
 
 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution | | Descriptor: | 11-cyclohexylundecanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 11-cyclohexylundecanoic acid and N- acetylglucosamine at 2.30 A resolution
To be Published
|
|
4OEK
 
 | | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYLETHYLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kumar, M, Singh, R.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of goat Lactoperoxidase with Phenylethylamine at 2.47 A
To be Published
|
|
4OEI
 
 | | Crystal structure of plant lectin from Cicer arietinum at 2.6 angstrom resolution | | Descriptor: | Lectin, MAGNESIUM ION, SULFATE ION | | Authors: | Kumar, S, Dube, D, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2014-01-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of plant lectin from Cicer arietinum at
2.6 angstrom resolution
TO BE PUBLISHED
|
|
4ORV
 
 | | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-phenylheptanoic acid, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of camel peptidoglycan recognition protein PGRP-S with 7- phenylheptanoic acid and N- acetylglucosamine at 2.50 A resolution
To be Published
|
|
3LPU
 
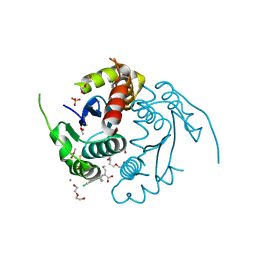 | | HIV integrase | | Descriptor: | (2S)-2-(6-chloro-2-methyl-4-phenylquinolin-3-yl)pentanoic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-06 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
3LPT
 
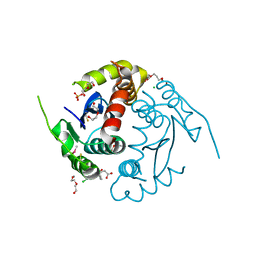 | | HIV integrase | | Descriptor: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
4GQB
 
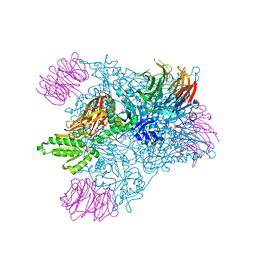 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6JKX
 
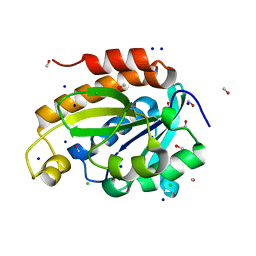 | | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution. | | Descriptor: | CHLORIDE ION, METHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution.
To Be Published
|
|
4U67
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 | | Descriptor: | 23s RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KZN
 
 | | Metabotropic Glutamate Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
7E72
 
 | | Crystal structure of Tie2-agonistic antibody in complex with human Tie2 Fn2-3 | | Descriptor: | 1,2-ETHANEDIOL, Angiopoietin-1 receptor, the chimeric Fab fragment of 3H7 (heavy chain), ... | | Authors: | Kim, H.M, Jo, G.H, Hong, H.J, Han, A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural insights into the clustering and activation of Tie2 receptor mediated by Tie2 agonistic antibody.
Nat Commun, 12, 2021
|
|
3V2K
 
 | | Crystal structure of ribosome inactivating protein from momordica balsamina complexed with the product of RNA substrate adenosine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
3MU6
 
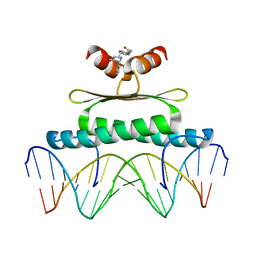 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
7VTN
 
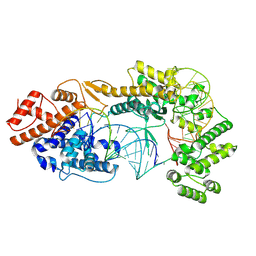 | | Cryo-EM structure of the Cas13bt3-crRNA-target RNA ternary complex | | Descriptor: | Cas13bt3, crRNA, target RNA | | Authors: | Nakagawa, R, Soumya, K, Han, A, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Tomohiro, N, Yamashita, K, Feng, Z, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-30 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
1R4K
 
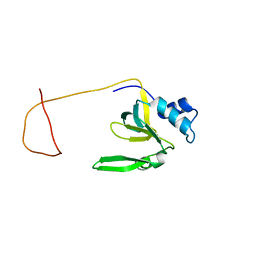 | | Solution Structure of the Drosophila Argonaute 1 PAZ Domain | | Descriptor: | Argonaute 1 | | Authors: | Yan, K.S, Yan, S, Farooq, A, Han, A, Zeng, L, Zhou, M.-M. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and conserved RNA binding of the PAZ domain
Nature, 426, 2003
|
|
2H8N
 
 | |
1LTZ
 
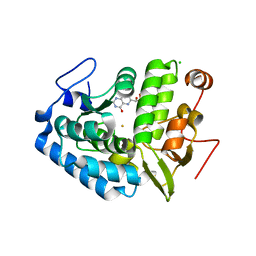 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-21 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | Descriptor: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | Deposit date: | 2002-05-20 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
