2NOJ
 
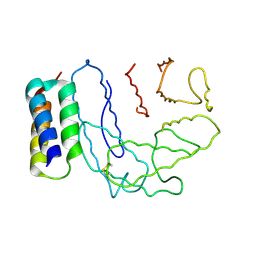 | | Crystal structure of Ehp / C3d complex | | Descriptor: | Complement C3, Efb homologous protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Ehp, a Secreted Complement Inhibitory Protein from Staphylococcus aureus.
J.Biol.Chem., 282, 2007
|
|
2GOM
 
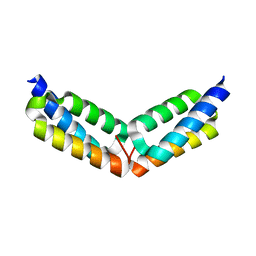 | |
2GOX
 
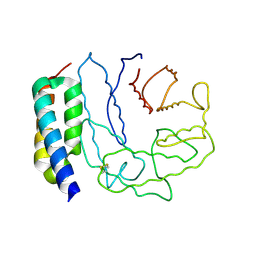 | | Crystal structure of Efb-C / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-04-14 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural basis for complement inhibition by Staphylococcus aureus.
Nat.Immunol., 8, 2007
|
|
3SR2
 
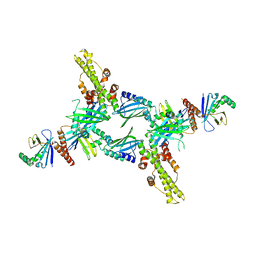 | | Crystal Structure of Human XLF-XRCC4 Complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Hammel, M, Classen, S, Tainer, J.A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.9708 Å) | | Cite: | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
4YFT
 
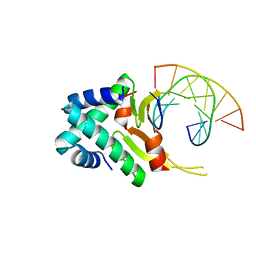 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEW
 
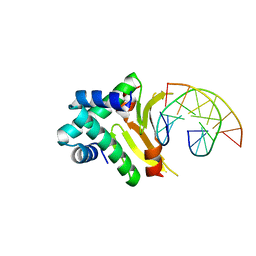 | | HUab-19bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.683 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEY
 
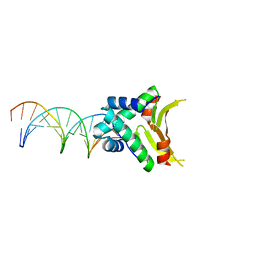 | | HUaa-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.354 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YFH
 
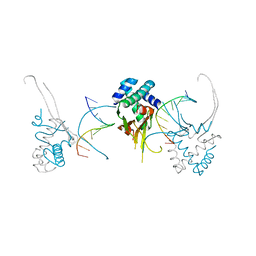 | | HU38-20bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YF0
 
 | | HU38-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
4YEX
 
 | | HUaa-19bp | | Descriptor: | DNA-binding protein HU-alpha, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-24 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
3L2P
 
 | | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching Between Two DNA Bound States | | Descriptor: | 5'-D(*GP*CP*CP*AP*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*CP*CP*G)-3', 5'-D(*GP*TP*CP*GP*GP*AP*CP*TP*G)-3', 5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*GP*TP*C)-3', ... | | Authors: | Cotner-Gohara, E.A, Kim, I.K, Hammel, M, Tainer, J.A, Tomkinson, A, Ellenberger, T. | | Deposit date: | 2009-12-15 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human DNA Ligase III Recognizes DNA Ends by Dynamic Switching between Two DNA-Bound States.
Biochemistry, 49, 2010
|
|
5UQY
 
 | | Crystal structure of Marburg virus GP in complex with the human survivor antibody MR78 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENVELOPE GLYCOPROTEIN GP1, ... | | Authors: | Hashiguchi, T, Fusco, M.L, Hastie, K.M, Bomholdt, Z.A, Lee, J.E, Flyak, A.I, Matsuoka, R, Kohda, D, Yanagi, Y, Hammel, M, Crowe, J.E, Saphire, E.O. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for Marburg virus neutralization by a cross-reactive human antibody.
Cell, 160, 2015
|
|
7TZ1
 
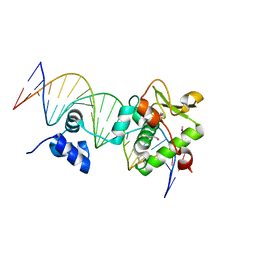 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
6O8Q
 
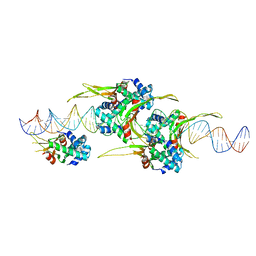 | | HUaa 19bp SYM DNA pH 4.5 | | Descriptor: | DNA (57-MER), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.216 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
7R6R
 
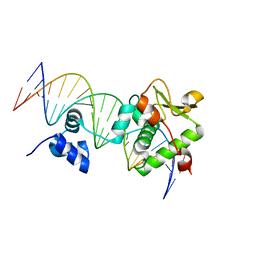 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
3DQV
 
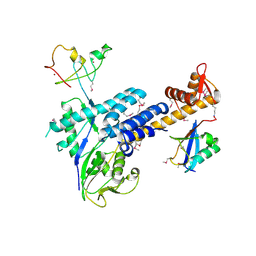 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | Descriptor: | Cullin-5, NEDD8, Rbx1, ... | | Authors: | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
6P7E
 
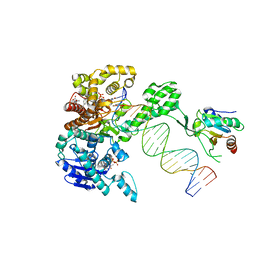 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
6OAJ
 
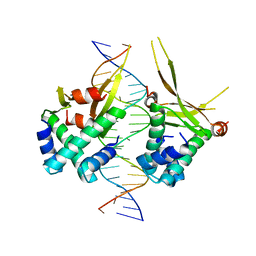 | | HUaE34K 19bp SYM DNA | | Descriptor: | DNA (5'-D(P*CP*GP*GP*TP*TP*CP*AP*AP*TP*TP*GP*GP*CP*AP*CP*GP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*CP*GP*TP*GP*CP*CP*AP*AP*TP*TP*GP*AP*AP*CP*CP*GP*C)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
6O6K
 
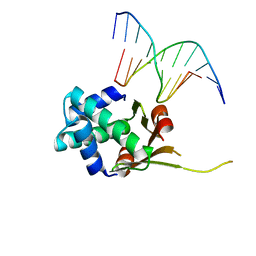 | | HUaa 19bp SYM DNA pH 5.5 | | Descriptor: | DNA (5'-D(P*AP*TP*TP*TP*CP*AP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*AP*TP*CP*AP*TP*GP*AP*AP*A)-3'), DNA-binding protein HU-alpha | | Authors: | Remesh, S.G, Hammel, M. | | Deposit date: | 2019-03-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.601 Å) | | Cite: | Nucleoid remodeling during environmental adaptation is regulated by HU-dependent DNA bundling.
Nat Commun, 11, 2020
|
|
3T7H
 
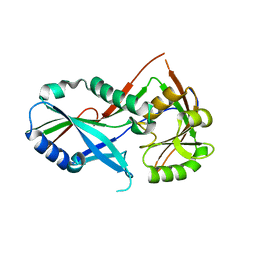 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7E
 
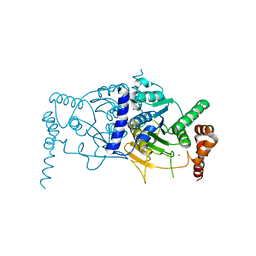 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7G
 
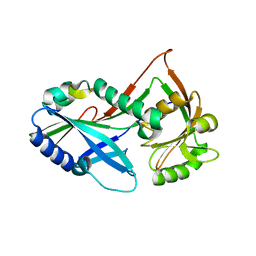 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Autophagy-related protein 3, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3T7F
 
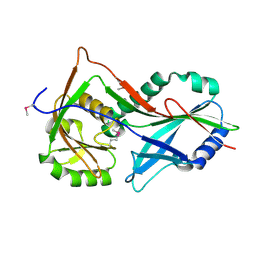 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
