7R45
 
 | | Bovine complex I in the presence of IM1761092, deactive class i (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R46
 
 | | Bovine complex I in the presence of IM1761092, deactive class ii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R43
 
 | | Bovine complex I in the presence of IM1761092, active class iii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R48
 
 | | Bovine complex I in the presence of IM1761092, deactive class iv (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R47
 
 | | Bovine complex I in the presence of IM1761092, deactive class iii (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R4C
 
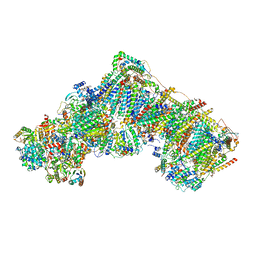 | | Bovine complex I in the presence of IM1761092, deactive class v (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
7R4F
 
 | | Bovine complex I in the presence of IM1761092, slack class i (Composite map) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-carbamimidoyl-3-[2-(3-chloranyl-4-iodanyl-phenyl)ethyl]guanidine, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-02-08 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
1GUW
 
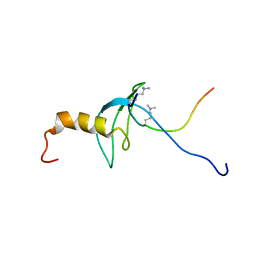 | | STRUCTURE OF THE CHROMODOMAIN FROM MOUSE HP1beta IN COMPLEX WITH THE LYSINE 9-METHYL HISTONE H3 N-TERMINAL PEPTIDE, NMR, 25 STRUCTURES | | Descriptor: | CHROMOBOX PROTEIN HOMOLOG 1, HISTONE H3.1 | | Authors: | Nielsen, P.R, Nietlispach, D, Mott, H.R, Callaghan, J.M, Bannister, A, Kouzarides, T, Murzin, A.G, Murzina, N.V, Laue, E.D. | | Deposit date: | 2002-02-01 | | Release date: | 2002-03-12 | | Last modified: | 2018-01-17 | | Method: | SOLUTION NMR | | Cite: | Structure of the Hp1 Chromodomain Bound to Histone H3 Methylated at Lysine 9
Nature, 416, 2002
|
|
1GRU
 
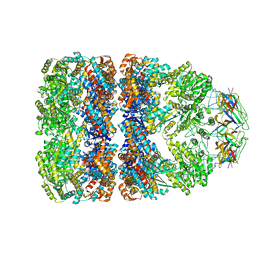 | | SOLUTION STRUCTURE OF GROES-ADP7-GROEL-ATP7 COMPLEX BY CRYO-EM | | Descriptor: | GROEL, GROES | | Authors: | Ranson, N.A, Farr, G.W, Roseman, A.M, Gowen, B, Fenton, W.A, Horwich, A.L, Saibil, H.R. | | Deposit date: | 2001-12-16 | | Release date: | 2002-01-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | ATP-Bound States of Groel Captured by Cryo-Electron Microscopy.
Cell(Cambridge,Mass.), 107, 2001
|
|
1P6U
 
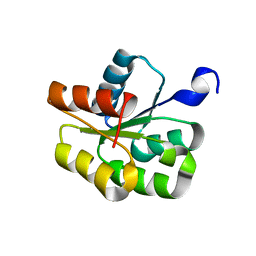 | | NMR structure of the BeF3-activated structure of the response regulator Chey2-Mg2+ from Sinorhizobium meliloti | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the Inactive and BeF(3)-activated Response Regulator CheY2
J.Biol.Chem., 338, 2004
|
|
7QIG
 
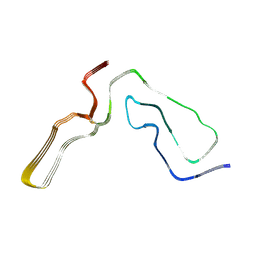 | | Infectious mouse-adapted RML scrapie prion fibril purified from terminally-infected mouse brains | | Descriptor: | Major prion protein | | Authors: | Manka, S.W, Zhang, W, Wenborn, A, Betts, J, Joiner, S, Saibil, H.R, Collinge, J, Wadsworth, J.D.F. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | 2.7 angstrom cryo-EM structure of ex vivo RML prion fibrils.
Nat Commun, 13, 2022
|
|
7QSD
 
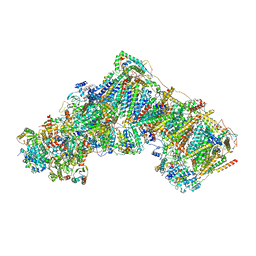 | | Bovine complex I in the active state at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
1HXN
 
 | |
1P6Q
 
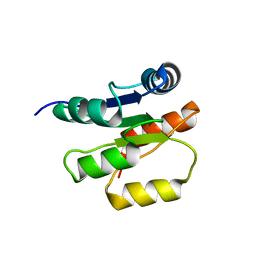 | | NMR Structure of the Response regulator CheY2 from Sinorhizobium meliloti, complexed with Mg++ | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the inactive and BeF3-activated response regulator CheY2.
J.Mol.Biol., 338, 2004
|
|
1HK6
 
 | | Ral binding domain from Sec5 | | Descriptor: | EXOCYST COMPLEX COMPONENT SEC5 | | Authors: | Mott, H.R, Nietlispach, D, Hopkins, L.J, Mirey, G, Camonis, J.H, Owen, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-03-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site.
J. Biol. Chem., 278, 2003
|
|
1OSH
 
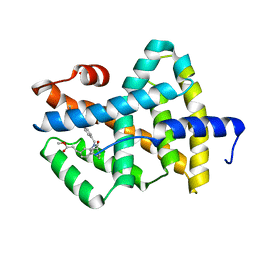 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
1FAR
 
 | |
1IRL
 
 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | Descriptor: | INTERLEUKIN-2 | | Authors: | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
1G6P
 
 | | SOLUTION NMR STRUCTURE OF THE COLD SHOCK PROTEIN FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | COLD SHOCK PROTEIN TMCSP | | Authors: | Kremer, W, Schuler, B, Harrieder, S, Geyer, M, Gronwald, W, Welker, C, Jaenicke, R, Kalbitzer, H.R. | | Deposit date: | 2000-11-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the cold-shock protein from the hyperthermophilic bacterium Thermotoga maritima.
Eur.J.Biochem., 268, 2001
|
|
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-10-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
1S4Z
 
 | | HP1 chromo shadow domain in complex with PXVXL motif of CAF-1 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromobox protein homolog 1 | | Authors: | Thiru, A, Nietlispach, D, Mott, H.R, Okuwaki, M, Lyon, D, Nielsen, P.R, Hirshberg, M, Verreault, A, Murzina, N.V, Laue, E.D. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin.
Embo J., 23, 2004
|
|
1SI5
 
 | | Protease-like domain from 2-chain hepatocyte growth factor | | Descriptor: | hepatocyte growth factor | | Authors: | Kirchhofer, D, Yao, X, Peek, M, Eigenbrot, C, Lipari, M.T, Billeci, K.L, Maun, H.R, Moran, P, Santell, L, Lazarus, R.A. | | Deposit date: | 2004-02-27 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling
J.Biol.Chem., 279, 2004
|
|
1JB9
 
 | | Crystal Structure of The Ferredoxin:NADP+ Reductase From Maize Root AT 1.7 Angstroms | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, ferredoxin-NADP reductase | | Authors: | Faber, H.R, Karplus, P.A, Aliverti, A, Ferioli, C, Spinola, M. | | Deposit date: | 2001-06-03 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and crystallographic characterization of ferredoxin-NADP(+) reductase from nonphotosynthetic tissues.
Biochemistry, 40, 2001
|
|
1GLA
 
 | | STRUCTURE OF THE REGULATORY COMPLEX OF ESCHERICHIA COLI IIIGLC WITH GLYCEROL KINASE | | Descriptor: | GLUCOSE-SPECIFIC PROTEIN IIIGlc, GLYCEROL, GLYCEROL KINASE | | Authors: | Hurley, J.H, Worthylake, D, Faber, H.R, Meadow, N.D, Roseman, S, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1992-10-28 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the regulatory complex of Escherichia coli IIIGlc with glycerol kinase.
Science, 259, 1993
|
|
1Q7X
 
 | | Solution structure of the alternatively spliced PDZ2 domain (PDZ2b) of PTP-Bas (hPTP1E) | | Descriptor: | PDZ2b domain of PTP-Bas (hPTP1E) | | Authors: | Kachel, N, Erdmann, K.S, Kremer, W, Wolff, P, Gronwald, W, Heumann, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-08-20 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing induced modulation of ligand specificity.
J.Mol.Biol., 334, 2003
|
|
