6J2S
 
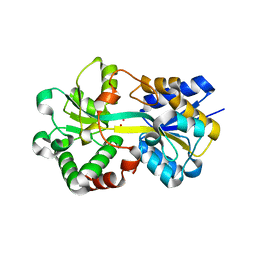 | | Structure of HitA bound to gallium from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding periplasmic protein, GALLIUM (III) ION, PHOSPHATE ION | | Authors: | Guo, Y, Li, H.Y, Sun, H.Z, Xia, W. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73005116 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
7VIB
 
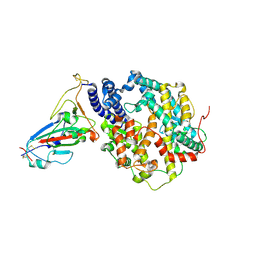 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
7XGC
 
 | |
7XGQ
 
 | |
3S0Z
 
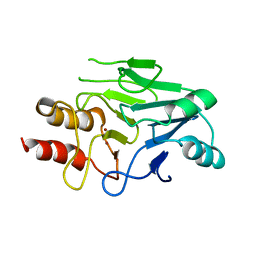 | | Crystal structure of New Delhi Metallo-beta-lactamase (NDM-1) | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Guo, Y, Wang, J, Niu, G.J, Shui, W.Q, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural view of the antibiotic degradation enzyme NDM-1 from a superbug.
Protein Cell, 2011
|
|
5Y6J
 
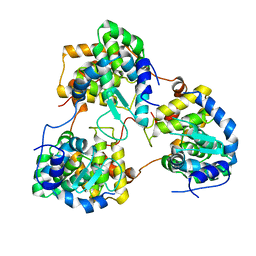 | |
8WEM
 
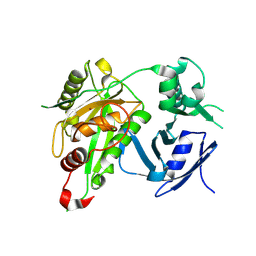 | | Bacteroides fragilis toxin 1 | | Descriptor: | Fragilysin, ZINC ION | | Authors: | Guo, Y, Wen, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteroides fragilis toxin 1
To Be Published
|
|
7VMU
 
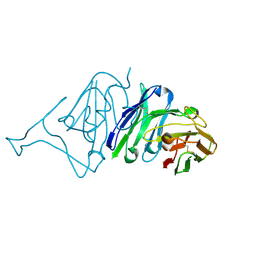 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | Spike protein S1, scFv E4 | | Authors: | Guo, Y, Wang, W, Jiao, P, Yang, H, Rao, Z, Cheng, G. | | Deposit date: | 2021-10-09 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Antibody engineering improves neutralization activity against K417 spike mutant SARS-CoV-2 variants.
Cell Biosci, 12, 2022
|
|
3U3I
 
 | | A RNA binding protein from Crimean-Congo hemorrhagic fever virus | | Descriptor: | Nucleocapsid protein | | Authors: | Guo, Y, Wang, W.M, Ji, W, Deng, M, Sun, Y.N, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2011-10-06 | | Release date: | 2012-03-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ZPW
 
 | | Generation of a long-acting fusion inhibitor against HIV-1 | | Descriptor: | MET-THR-TRP-GLU-GLU-TRP-ASP-MK8-LYS-ILE-GLU-MK8-TYR-THR-MK8-LYS-ILE-GLU-MK8-LEU-ILE-LYS-LYS-SER, Transmembrane protein gp41 | | Authors: | Guo, Y, Shi, X.L. | | Deposit date: | 2018-04-16 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Generation of a long-acting fusion inhibitor against HIV-1.
Medchemcomm, 9, 2018
|
|
7WWM
 
 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7WWL
 
 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7CJT
 
 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | Descriptor: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CJU
 
 | |
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
7CAJ
 
 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E21
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state with ATP-gamma-S | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E1Z
 
 | | Cryo EM structure of a Na+-bound Na+,K+-ATPase in the E1 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7E20
 
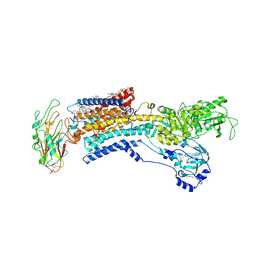 | | Cryo EM structure of a K+-bound Na+,K+-ATPase in the E2 state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Yan, R.H, Huang, B.D, Ye, F.F, Wu, L.S, Chi, X.M, Zhou, Q. | | Deposit date: | 2021-02-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states.
Nat Commun, 13, 2022
|
|
7C9N
 
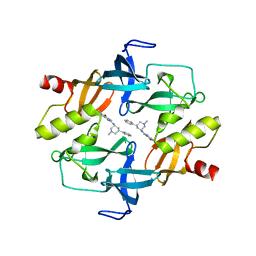 | | Crystal structure of SETDB1 tudor domain in complexed with Compound 1. | | Descriptor: | 3,5-dimethyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | Authors: | Guo, Y, Xiong, L, Mao, X, Yang, S. | | Deposit date: | 2020-06-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DOQ
 
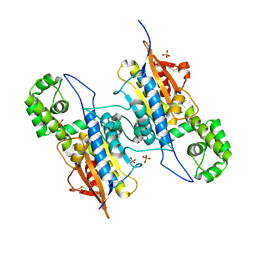 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | Acid phosphatase, PHOSPHATE ION | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-12-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7D2F
 
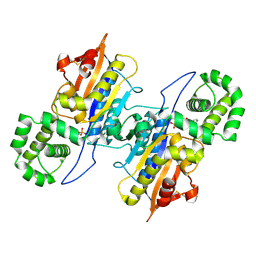 | | Lp major histidine acid phosphatase mutant D281A/5'-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Major acid phosphatase | | Authors: | Guo, Y, Teng, Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into a new substrate binding mode of a histidine acid phosphatase from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
7CZA
 
 | |
