3N4G
 
 | | Crystal structure of native Cg10062 | | Descriptor: | Putative tautomerase | | Authors: | Guo, Y, Robertson, B.A, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-05-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of the Native and Inactivated Cg10062, a cis-3-Chloroacrylic Acid Dehalogenase from Corynebacterium glutamicum: Implications for the Evolution of cis-3-Chloroacrylic Acid Dehalogenase Activity in the Tautomerase Superfamily
To be Published
|
|
3N4D
 
 | |
3MQ3
 
 | |
3MJZ
 
 | | The crystal structure of native FG41 MSAD | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson, W.H.Jr, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-13 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
4LPZ
 
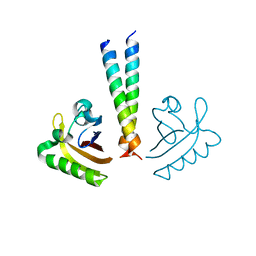 | | ARNT transcription factor/coactivator complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Transforming acidic coiled-coil-containing protein 3 | | Authors: | Guo, Y, Scheuermann, T.H, Gardner, K.H. | | Deposit date: | 2013-07-16 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Coiled-coil coactivators play a structural role mediating interactions in hypoxia-inducible factor heterodimerization.
J.Biol.Chem., 290, 2015
|
|
4LHP
 
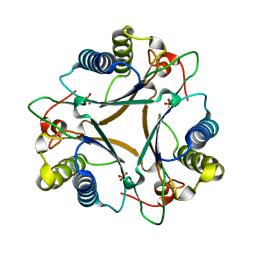 | | Crystal Structure of Native FG41Malonate Semialdehyde Decarboxylase | | Descriptor: | FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MF7
 
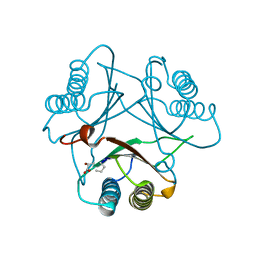 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
7L97
 
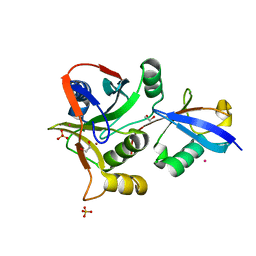 | | Crystal structure of STAMBPL1 in complex with an engineered binder | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease, SULFATE ION, ... | | Authors: | Guo, Y, Dong, A, Hou, F, Li, Y, Zhang, W, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-01-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.
J.Biol.Chem., 297, 2021
|
|
6LR1
 
 | |
3N4H
 
 | |
3F70
 
 | | Crystal structure of L3MBTL2-H4K20me1 complex | | Descriptor: | Lethal(3)malignant brain tumor-like 2 protein, N-METHYL-LYSINE | | Authors: | Guo, Y, Qi, C, Allali-Hassani, A, Pan, P, Zhu, H, Dong, A, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Edwards, A.M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Botchkarev, A, Read, R, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-07 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2.
Nucleic Acids Res., 37, 2009
|
|
4LHO
 
 | | Crystal Structure of FG41Malonate Semialdehyde Decarboxylase inhibited by 3-bromopropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase, PHOSPHATE ION | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
3MF8
 
 | | Crystal Structure of Native cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase, SULFATE ION | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
3MLC
 
 | | Crystal structure of FG41MSAD inactivated by 3-chloropropiolate | | Descriptor: | 3-chloro-3-oxopropanoic acid, FG41 Malonate Semialdehyde Decarboxylase | | Authors: | Guo, Y, Serrano, H, Poelarends, G.J, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Kinetic, Mutational, and Structural Analysis of Malonate Semialdehyde Decarboxylase from Coryneform Bacterium Strain FG41: Mechanistic Implications for the Decarboxylase and Hydratase Activities.
Biochemistry, 52, 2013
|
|
5F5O
 
 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
6B3R
 
 | | Structure of the mechanosensitive channel Piezo1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1, unknown fragment | | Authors: | Guo, Y.R, MacKinnon, R. | | Deposit date: | 2017-09-22 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-based membrane dome mechanism for Piezo mechanosensitivity.
Elife, 6, 2017
|
|
5E04
 
 | | Crystal structure of Andes virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W.M, Lou, Z.Y. | | Deposit date: | 2015-09-28 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Core Region of Hantavirus Nucleocapsid Protein Reveals the Mechanism for Ribonucleoprotein Complex Formation
J.Virol., 90, 2015
|
|
6IWF
 
 | | Crystal structure of HitA from Pseudomonas aeruginosa | | Descriptor: | Ferric iron-binding protein HitA | | Authors: | Guo, Y, Zhengrui, Z, Li, H. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.70662332 Å) | | Cite: | Identification and Characterization of a Metalloprotein Involved in Gallium Internalization in Pseudomonas aeruginosa.
Acs Infect Dis., 5, 2019
|
|
5E06
 
 | |
5E05
 
 | |
5F5M
 
 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
6HNQ
 
 | | TarP-6RboP-(CH2)6NH2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable ss-1,3-N-acetylglucosaminyltransferase, ... | | Authors: | Guo, Y, Stehle, T. | | Deposit date: | 2018-09-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus alters cell wall glycosylation to evade immunity.
Nature, 563, 2018
|
|
8WEN
 
 | | Bacteroides fragilis toxin 2 | | Descriptor: | Metalloprotease, SODIUM ION, ZINC ION | | Authors: | Guo, Y, Wen, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Bacteroides fragilis toxin 2
To Be Published
|
|
8WEO
 
 | |
3U1U
 
 | | Crystal structure of RNA polymerase-associated protein RTF1 homolog Plus-3 domain | | Descriptor: | GLYCEROL, RNA polymerase-associated protein RTF1 homolog, SULFATE ION, ... | | Authors: | Guo, Y, Tempel, W, Bian, C, Wernimont, A.K, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2012-04-04 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of RNA polymerase-associated protein RTF1 homolog Plus-3 domain
to be published
|
|
