4KDV
 
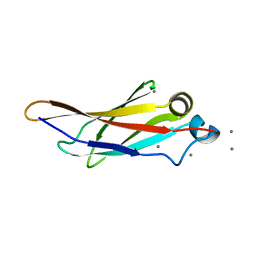 | | Crystal structure of a bacterial immunoglobulin-like domain from the M. primoryensis ice-binding adhesin | | Descriptor: | Antifreeze protein, CALCIUM ION | | Authors: | Guo, S, Garnham, C.P, Karunan, S.P, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Role of Ca(2+) in folding the tandem beta-sandwich extender domains of a bacterial ice-binding adhesin.
Febs J., 280, 2013
|
|
4KDW
 
 | | Crystal structure of a bacterial immunoglobulin-like domain from the M. primoryensis ice-binding adhesin | | Descriptor: | Antifreeze protein, CALCIUM ION, GLYCEROL | | Authors: | Guo, S, Garnham, C.P, Karunan, S.P, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Role of Ca(2+) in folding the tandem beta-sandwich extender domains of a bacterial ice-binding adhesin.
Febs J., 280, 2013
|
|
5IRB
 
 | | Structural insight into host cell surface retention of a 1.5-MDa bacterial ice-binding adhesin | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Guo, S, Phippen, S, Campbell, R, Davies, P. | | Deposit date: | 2016-03-12 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
8XJ3
 
 | |
1CR0
 
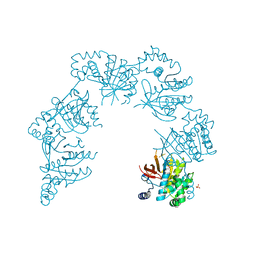 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE4 PROTEIN OF BACTERIOPHAGE T7 | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1I9Y
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN | | Descriptor: | PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | Authors: | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | Deposit date: | 2001-03-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I9Z
 
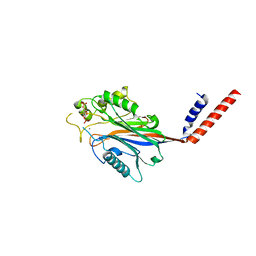 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN IN COMPLEX WITH INOSITOL (1,4)-BISPHOSPHATE AND CALCIUM ION | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | Authors: | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | Deposit date: | 2001-03-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
8YGW
 
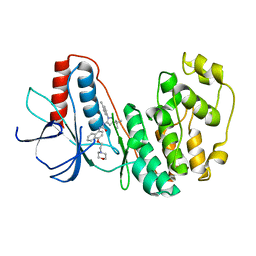 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
8EF5
 
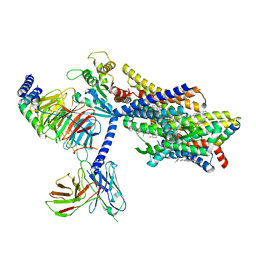 | | Fentanyl-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EF6
 
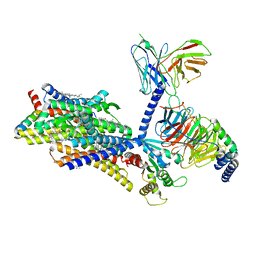 | | Morphine-bound mu-opioid receptor-Gi complex | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFO
 
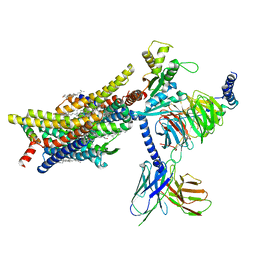 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFL
 
 | | SR17018-bound mu-opioid receptor-Gi complex | | Descriptor: | 5,6-dichloro-1-{1-[(4-chlorophenyl)methyl]piperidin-4-yl}-1,3-dihydro-2H-benzimidazol-2-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFB
 
 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
1CR1
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR2
 
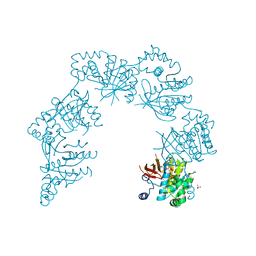 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
6DDA
 
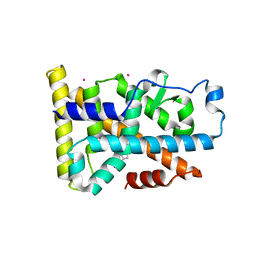 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
9JFB
 
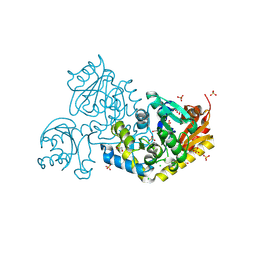 | | Crystal structure of L-threonine-O-3-phosphate decarboxylase CobC | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Jiang, M, Guo, S, Chen, X, Wei, Q, Wang, M. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of l-threonine-O-3-phosphate decarboxylase CobC from Sinorhizobium meliloti involved in vitamin B 12 biosynthesis.
Biochem.Biophys.Res.Commun., 734, 2024
|
|
9JFF
 
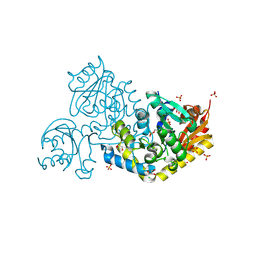 | | Crystal structure of L-threonine-O-3-phosphate decarboxylase CobC in complex with reaction intermediate | | Descriptor: | CHLORIDE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Jiang, M, Guo, S, Chen, X, Wei, Q, Wang, M. | | Deposit date: | 2024-09-04 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of l-threonine-O-3-phosphate decarboxylase CobC from Sinorhizobium meliloti involved in vitamin B 12 biosynthesis.
Biochem.Biophys.Res.Commun., 734, 2024
|
|
4PES
 
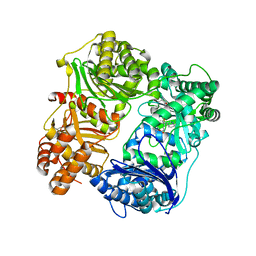 | |
4PF9
 
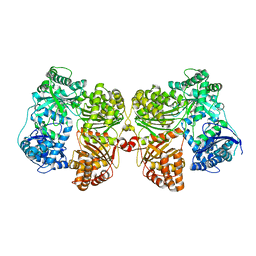 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-[4-({5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}oxy)phenyl]-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
4PF7
 
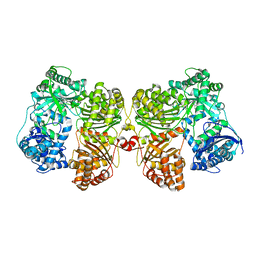 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
4PFC
 
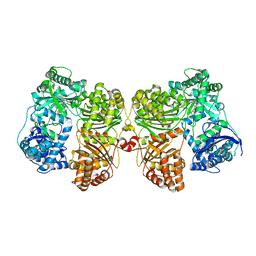 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl [(2S)-2-(5-{5-[4-({(2S)-2-[(3S)-3-amino-2-oxopiperidin-1-yl]-2-cyclohexylacetyl}amino)phenyl]pentyl}-2-fluorophenyl)-3-(quinolin-3-yl)propyl]carbamate | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
3JU5
 
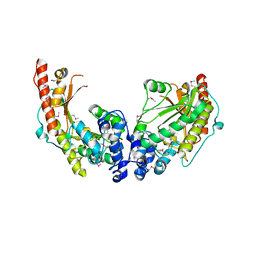 | | Crystal Structure of Dimeric Arginine Kinase at 1.75-A Resolution | | Descriptor: | Arginine kinase, MAGNESIUM ION | | Authors: | Wu, X, Ye, S, Guo, S, Yan, W, Bartlam, M, Rao, Z. | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for a reciprocating mechanism of negative cooperativity in dimeric phosphagen kinase activity
Faseb J., 24, 2010
|
|
